
SHIC, launched in 2015 with Pork Checkoff funding, continues to focus efforts on prevention, preparedness, and response to novel and emerging swine disease for the benefit of US swine health. As a conduit of information and research, SHIC encourages sharing of its publications and research. Forward, reprint, and quote SHIC material freely. SHIC is funded by America’s pork producers to fulfill its mission to protect and enhance the health of the US swine herd. For more information, visit http://www.swinehealth.org or contact Dr. Sundberg at [email protected].
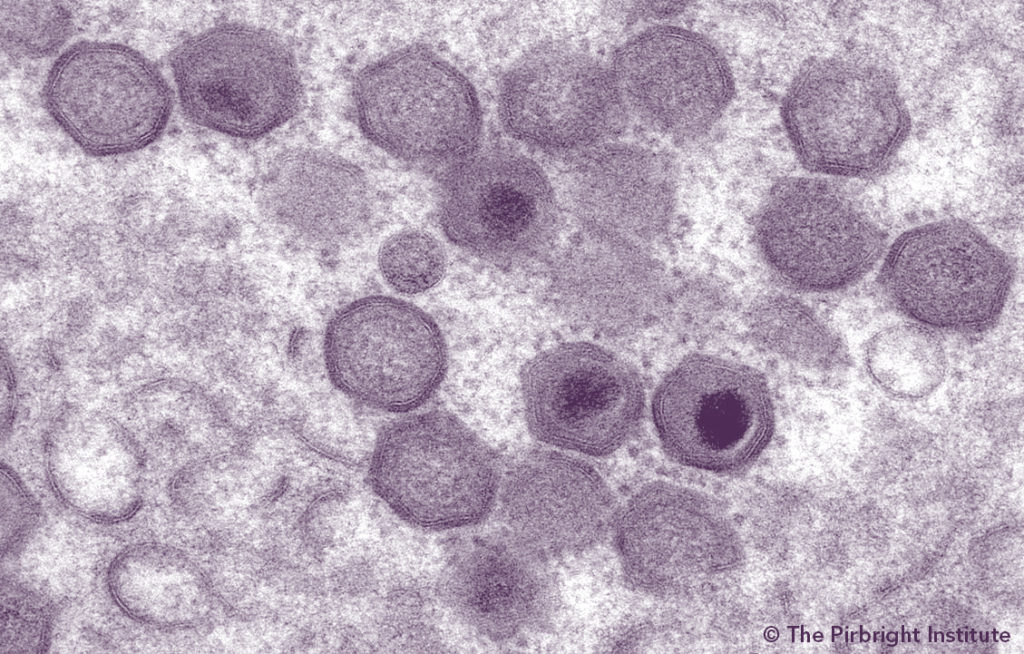
A project to summarize current knowledge and remaining gaps regarding the molecular epidemiology of African swine fever (ASF) was funded by the Swine Health Information Center (SHIC). The unprecedented expansion of ASF during the last five years produced a 218% increase in the volume of scientific publications on the subject compared to the previous five years (2010 to 2014). Due to this volume, a critical need to synthesize available scientific evidence to support and facilitate translation of the new evidence into updates for regulations and policy framework, and management recommendations for the industry, was apparent. Staff at the Center for Animal Health and Food Safety (CAHFS) at the University of Minnesota College of Veterinary Medicine conducted a systematic review of current literature including, but not restricted to, genetic diversity of strains, the association between sequence data, and epidemiological or pathogenic features, and development and performance of molecular diagnostic tools.
Resulting information from this study will inform appropriate preparedness and control strategies in the US. This work found globally available data, and consequently, analytical approaches, show a strong asymmetry across genotypes. This gap demonstrates eastern and southern Africa’s role as a major reservoir of virus diversity, from which new spillovers such as the one in 2007 could happen again, which needs to be continually addressed. It also reported several comprehensive lists of research priorities to be addressed to support more efficient control strategies on a global scale (Table 1). Identifying relevant research gaps in disease prevention and control informs the ultimate goal of supporting experiments and projects to fill those gaps.
GENOMICS GAPS:
MOLECULAR DIAGNOSIS & SURVEILLANCE GAPS:

A research project on African swine fever (ASF) in Vietnam, made possible by a USDA Foreign Ag Service grant obtained by the Swine Health Information Center (SHIC) with support from the National Pork Producers Council, examined the potential for rodents to serve as vectors of the devastating virus. Rodent vectors are a possible transmission route long-established for other swine diseases, but uncharacterized for ASF. Work conducted by researchers from South Dakota State University and the Vietnam National University of Agriculture (VNUA) on Vietnamese farms with differing biosecurity levels provided information that suggests rodents are not a high risk of being ASF vectors.
One objective of the study was to determine whether rodents trapped in and around ASF-infected farms harbored ASFV (African swine fever virus), and if so, the optimal samples to obtain from rodents to detect viral infection. Five commercial farms located across four provinces in northern Vietnam with recent ASF outbreaks were identified. The farms ranged in size from 26 to 851 pigs. Biosecurity procedures varied among the farms; two farms featured closed housing, one open housing, and two combined closed and open housing.
Live traps were placed nightly at each study farm, outside facilities at entry points, and inside facilities near feed sources. Trapping continued at each farm for 10 to 36 days. Rats were euthanized on the farm, stored in freezers, and transported to VNUA for necropsy and PCR testing.
Samples from rats in and around farms undergoing active ASF outbreaks were negative for ASF across several different sample types (examining biological as well as mechanical vector potential). This work suggests that rats (and presumably similar rodents) do not serve as important vectors for ASFV.
Building on the negative results from Experiment 1, the second experiment examined whether rats were susceptible to challenge with ASFV, and if so whether they were able to transmit the virus to susceptible rodents.
Rats were randomly assigned to one of six ASF challenge cages. In each challenge cage, rats were challenged with a dose of 105 50% hemadsorption doses (HAD50) ASF either intraperitoneally or orally, while additional rats in each cage served as susceptible contacts. One cage of rats served as non-inoculated controls. Body temperatures and clinical signs were recorded three times daily for each animal.
Clinical signs were not present in any rats during the observation periods. Initial analysis indicates there were no differences between the body temperature of control and inoculated rats, although temperatures of all rats (control, inoculated, and contacts) tended to climb during the second week of the experiment, then fall back to baseline. Despite robust challenges both intraperitoneally and orally, rats were not observed to become ill from nor infected with ASFV out to an incubation period of 21 days.
This characterization provides insight for biosecurity protocols surrounding prevention of ASF outbreaks. While rodents do not appear to be a significant risk as ASF vectors, other work has proven them to be disease carriers and appropriate biosecurity protocols for elimination continue to be recommended.

In 2019, a new variant of porcine sapovirus (PSaV) infection was identified in the US on a farm reporting piglet diarrhea issues for two years. Piglet SaV infection during the lactation phase resulted in smaller pigs at weaning, losing one to two pounds, which represents a significant health and economic consequence. PSaV is included on SHIC’s Swine Viral Disease Matrix; knowing the epidemiology and having effective diagnostics for those pathogens is key to effective management.
The Swine Health Information Center (SHIC), funded by Pork Checkoff, worked with Iowa State University researchers to help understand PSaV’s role in causing disease and to develop appropriate diagnostics as part of its ongoing mission to monitor and respond to emerging swine diseases. The full study report is now available as well as a highly sensitive and specific RT-PCR for detecting porcine sapovirus (PSaV) genotype III for neonatal diarrhea investigation.
As part of the study, researchers used four independent lines of evidence – metagenomics analysis, real-time RT-PCR, histopathology, and in situ hybridization – to confirm that porcine PSaV of genogroup III as the cause of the enteritis and diarrhea on the farm. A subsequent prevalence survey of more than 500 samples comparing pigs with clinical diarrhea and clinically healthy pigs suggests that porcine SaV genotype III may play an important role in causing swine enteritis and diarrhea. Study findings provide significant insights for a better understanding of the epidemiology and pathogenicity of porcine PSaV. To the researchers’ knowledge, this is the first evidence PSaV likely serves as the sole etiological agent causing enteritis and diarrhea of piglets in the field in the US.
Porcine SaVs are genetically diverse and widely distributed in pig-producing countries. To date, eight genogroups of porcine SaV have been identified, and genogroup III (GIII) is the predominant type. Most of the eight genogroups of porcine SaV are circulating in the US. In the study, reports on detection of porcine SaVs in pigs at different ages with clinical diarrhea using next-generation sequencing and genetic characterization are included.
All seven cases in the study had porcine SaV GIII strains detected and one pooled case was found to have a porcine SaV GVI strain IA27912-B-2018, per the final report. Sequence analysis showed that seven GIII isolates were genetically divergent and formed four different lineages on the trees of complete genome, RdRP, VP1 and VP2. In addition, these seven GIII isolates had three different deletion/insertion patterns in an identified variable region close to the three prime end of VP2. The GVI strain IA27912-B-2018 was closely related to strains previously detected in the US and Japan. A 3-nt deletion in VP1 region of GVI IA27912-B-2018 was identified. The study showed genetically divergent PSaVs of different genogroups are co-circulating in pigs in the US.
Study authors conclude future studies comparing the virulence of these different genogroups in pigs are needed to better understand this virus. Developing new diagnostic tools will also help with understanding the incidence of PSaV and its contribution to piglet diarrheatic syndromes.
Pseudorabies virus (PRV) is ranked fourth in the Swine Health Information Center (SHIC) Swine Disease Matrix in large part due to the potential for the introduction of highly pathogenic PRV into the US from Asia – an event which would have a highly negative impact on pork exports. In addition, while PRV was officially eliminated from US domestic swine in 2004, it is occasionally introduced into “transitional” herds via contact with feral swine. For these reasons, improvements in PRV diagnostics, surveillance, control, and elimination remain relevant. The final report on a study conducted at Iowa State University (ISU) to evaluate the detection of PRV in swine oral fluid, providing additional testing resources using real-time PCR assays, has been posted. This research involving PCR PRV oral fluid testing was supported by a grant from SHIC.
Widely used PRV PCRs were developed for individual animal nasal swabs, but not for swine oral fluid. Therefore, ISU researchers set out to evaluate the detection of PRV in swine oral fluid collected from vaccinated and/or inoculated pigs using a contemporary real-time PCR assay targeting PRV gB genes. The results showed that PRV DNA could be detected in swine oral fluid specimens using a PRV gB PCR. Based on comparisons of detection rates in nasal swab vs oral fluid samples, the researchers also found that oral fluid could be used as an alternative to individual pig (nasal swab) sampling for PRV surveillance. They cautioned that further improvements in the performance of the gB PCRs would be recommended before its use in routine surveillance.
A separate study conducted at ISU and published in 2020 addressed the detection of PRV antibody in oral fluids. Analyses of an oral fluid PRV indirect IgG ELISA showed good binary diagnostic performance and excellent assay repeatability. Importantly, this study demonstrated the feasibility of detecting PRV antibodies in oral fluid specimens. This represents the first step toward the development of a DIVA-compatible oral fluid–based ELISA for use in PRV control and elimination programs.
Together, these two bodies of work illustrate the progress being made to prevent and respond to potential PRV outbreaks in the US swine herd.

The Swine Health Information Center (SHIC) has updated its Fact Sheet on Getah virus (GETV) and created a new one on Salmonella 1,4,[5],12:i:-, an emerging serotype in swine. The updated GETV Fact Sheet contains details about this viral disease gleaned from a comprehensive literature review of the latest research. Salmonella 1,4,[5],12:i:- has become one of the most identified serotypes in pigs, pork, and humans worldwide. It captured the attention of an American Association of Swine Veterinarians member who recommended the Swine Health Information Center (SHIC) develop a fact sheet on this bacteria for the benefit of the US pork industry.
Fact Sheet updates and additions represent SHIC’s mission of responding to industry needs. You are welcome to send ideas, questions, and suggestions for SHIC Fact Sheets by email, [email protected], or call, 515-598-4553. SHIC will provide a timely response with good science to benefit the industry, maintaining a regularly updated Fact Sheet library.
In the improved literature review portion of the Fact Sheet, in addition to sections on etiology, cleaning and disinfection, epidemiology, transmission, pathogenesis, diagnosis immunity, prevention, and gaps in preparedness, new sections on importance, public health, infection in swine, treatment, and history in swine have been added.
Additional information is incorporated as part of a Fact Sheet template update. Fact Sheets continue to begin with a summary section for easy access to the most important information. But the comprehensive literature review process and insertion of additional information into the fact sheets results in more complete information on the emerging swine disease. The order of those sections was changed to be more intuitive:
GETV Update
GETV is found throughout Eurasia. It has been mostly associated with outbreaks in horses, particularly in Japan. However, GETV is also known to cause disease in neonatal pigs and fetal death in pregnant sows. GETV mutates rapidly, and it is transmitted by different mosquito species whose distribution is ever changing. A better understanding of the epidemiology of GETV in pigs is needed to prepare for future outbreaks.
The main serotypes associated with clinical salmonellosis in pigs are S. typhimurium and S. 1,4,[5],12:i:-. Enterocolitis and septicemia can be seen. Most pigs recover but they may shed bacteria for months after resolution of clinical signs. Swine are also subclinical carriers of S. 1,4,[5],12:i:- and many other Salmonella serotypes. In carrier animals, shedding is exacerbated by stress including commingling, transport, and food deprivation.
Salmonella spp. can survive for long periods in the environment and are isolated from many sources.
For disinfection purposes, Salmonellae are generally susceptible to 1% sodium hypochlorite, 70% ethanol, 70% propanol, 2% glutaraldehyde, and 4% formaldehyde, as well as phenol, peracetic acid, hydrogen peroxide, quaternary ammonium compounds, and iodophors. A study of S. 1,4,[5],12:i:- in swine slaughterhouses found that isolates were susceptible to 0.5% peracetic acid, 1% peracetic acid, and 0.5% quaternary ammonium.
To prevent shedding in carrier animals, reduce environmental and transport-related stress. Additionally, pens and contaminated fomites must be cleaned and disinfected thoroughly to reduce Salmonella load in the environment. Producers should keep feed in rodent-proof containers to prevent contamination.
Antimicrobials are not indicated for treatment of subclinical salmonellosis. Most Salmonella transmission is fecal-oral. However, direct pig-to-pig transmission, inhalation, and
vertical transmission also occur.
To diagnose, culture and biochemical testing are used to identify Salmonella, but subtyping is necessary to determine serotype. Techniques used for subtyping include classical serotyping (White-Kauffman scheme), pulsed field gel electrophoresis (PFGE), multiple locus variable number of tandem repeats (MLVA), multilocus sequence typing (MLST), and whole genome sequencing (WGS).
PCR assays are not used for routine diagnosis, but several assays have been described that can differentiate S. 1,4,[5],12:i:- from S. typhimurium. ELISAs may be useful for herd-level screening, and there are several commercial assays available based on lipopolysaccharide (LPS) O-antigen that can identify Salmonella to the serogroup level. For pigs with diarrhea, pooled ileum, colon, and ileocecal lymph node are preferred for culture. Feces or tonsil scrapings can be collected from live pigs. In cases of septicemia, blood, lung, liver, and spleen are acceptable. Most ELISAs can be used with serum or meat juice. In addition, oral fluids may be useful as a surveillance tool.
Antimicrobial treatment is indicated for pigs with clinical signs and lesions suggestive of salmonellosis. Treatment regimens should be based on antibiograms, especially for emerging serotypes like S. 1,4,[5],12:i:- that often demonstrate multi-drug resistance.
Per the S. 1,4,[5],12:i:- Fact Sheet, nontyphoidal Salmonella are a leading cause of foodborne infections in humans. Animals are reservoirs for many salmonellae, including S. 1,4,[5],12:i:-, a monophasic variant of Salmonella typhimurium. Isolates are often resistant to multiple antimicrobials and heavy metals, making S. 1,4,[5],12:i:- a public health concern.
Further information on pathogenesis, epidemiology, etiology, and immunity, along with all references for data, are included in the S. 1,4,[5],12:i:- Fact Sheet.

The Swine Health Information Center (SHIC) 2021 Plan of Work includes the investigation of biocontainment and bioexclusion as tools to help prevent or control an emerging swine disease outbreak on farms. To achieve this objective, SHIC is soliciting proposals to investigate cost-effective, innovative technologies, protocols, or ideas to implement biocontainment in the face of an emerging disease outbreak on swine farms. SHIC is also soliciting proposals to investigate cost-effective, innovative technologies, protocols, or ideas to implement bioexclusion to prevent or control an emerging disease outbreak on swine farms.
Biocontainment, as defined in medical dictionaries, usually refers to procedures needed to contain pathogens in laboratories. The SHIC 2021 Plan of Work addresses biocontainment saying, “Decreasing the amount of pathogens from leaving an outbreak site will help to protect neighboring farms and regions from emerging disease outbreaks. Assessing existing technologies or new ideas for cost-effectiveness will help inform producers’ decisions about implementation.”
Effective biocontainment methodologies may be applied to a variety of viruses and/or bacteria, may be applied dependent upon expected transmission mode(s), or may be dependent on the epidemiology of the pathogen(s). Therefore, the biocontainment proposal should:
There is a pool of approximately $100,000 available for the biocontainment research, but individual project proposals could be higher with sufficient justification. Collaborative projects demonstrating the most urgency and timeliness of completion, as well as efficient use of funds, will be prioritized for funding. The proposal template and instructions for completion and submission can be found here. The deadline for proposal submission is 5:00 PM CDT, May 7, 2021.
Bioexclusion refers to procedures needed to prevent entry of a pathogen or disease and contact with a population of animal, a part of biosecurity. Specifically, SHIC would like proposals that address the following 2021 Plan of Work items:
There is a pool of approximately $150,000 available for the bioexclusion research, but individual project proposals could be higher with sufficient justification. Collaborative projects demonstrating the most urgency and timeliness of completion, as well as efficient use of funds, will be prioritized for funding. The proposal template and instructions for completion and submission can be found here. The deadline for proposal submission is 5:00 PM CDT, May 14, 2021.
For questions on these requests for research funding proposals, contact Dr. Paul Sundberg at [email protected] or (515) 451-6652.

SHIC Talk host Barb Determan visits with Swine Health Information Center (SHIC) Board President Dr. Daryl Olsen and SHIC Working Group member Dr. Joe Connor about the Center’s work since inception in 2015.
Hear their comments on the SHIC Program Review here.
As the world deals with the COVID-19 pandemic, SHIC continues to focus efforts on prevention, preparedness, and response to novel
and emerging swine disease for the benefit of US swine health.

This month’s Domestic Swine Disease Monitoring Report shows that porcine reproductive and respiratory syndrome virus (PRRSV) had a moderate increase in detection from March. Porcine delta coronavirus (PDCoV) activity observed since January is explained by increased testing following outbreaks observed in sow farms and viral activity in grow-finish animals. In the SDRS data, a site may have turned positive, kept submitting samples for testing, and obtained positive results over time but information on the incidence level was not known. Higher incidence is assumed because of the positivity in some regions. During the last year, since May 2020, only eight cases tested positive for transmissible gastroenteritis virus (TGEV). Mycoplasma hyopneumoniae was within expected levels for this time of year. At a state level, PRRSV detection was three standard deviations above expected in Nebraska, Ohio, and North Carolina; and PDCoV in Kansas, Oklahoma, Missouri, and North Carolina. In the podcast, the SDRS hosts talk with Dr. Eric Burrough about disease diagnosis coding and trends, and implications for researchers, diagnosticians, and practitioners.

Read this month’s Global Disease Monitoring Report to learn more about how, in collaboration with Zoetis Philippines, authorities have launched their first African swine fever (ASF) vaccine trials there. Read about Chinese authorities launching a regionalization strategy to control ASF. And understand how Australian border surveillance resulted in pork products testing positive for ASF and foot-and-mouth disease (FMD) being seized.
Copyright 2025 | Swinehealth.org | Website by Heartland Marketing Group