
Project #: 23-077 | Principal Investigator: Michael C. Rahe | Institution: North Carolina State University | Posted: 6/23/25 | Keywords: PoAstV4, tracheitis, bronchitis, CDCD pigs, machine learning, immunohistochemistry, in situ hybridization
Porcine astrovirus 4 (PoAstV4) has previously been detected in piglets with respiratory disease. However, it was unclear if the virus was the cause of disease or merely a bystander. The objective of this study was to infect piglets with PoAstV4 and evaluate if respiratory disease was reproduced. Caesarean-derived colostrum-deprived (CDCD) piglets were infected (17 – challenged and 11 negative controls) with PoAstV4 and were found to be shedding the virus in nasal secretions as early as 2 days post challenge (DPC) with all pigs testing negative by 14 DPC. Both tracheitis and bronchitis were diagnosed in infected pigs necropsied at 5 and 8 DPC with viral detection in these tissues at the same time. There was a productive immune response to infection with detection of anti-PoAstV4 antibodies in the blood and the characterization of immune cells within infected tissues. Results from this study show that PoAstV4 can cause microscopic lesions of an epitheliotropic viral infection in the respiratory tract, very similar to influenza A virus.
Project #: 23-068 | Principal Investigator: Igor Paploski and Cesar Corzo | Institution: University of Minnesota | Posted: 4/11/25 | Keywords: epidemiology, tongue tips, disease testing, surveillance, PRRSV, swine, infectious diseases
Porcine reproductive and respiratory syndrome virus (PRRSV) causes significant economic losses in the U.S., approximately USD 1.2 billion annually, due to reproductive failure, abortion, and high pre-weaning mortality among piglets. Approximately 30% of the U.S. breeding herd experiences a PRRSV outbreak every year. Tongue tips from dead animals, particularly piglets, are being considered as an alternative specimen to monitor PRRS during herd stabilization; however, questions as to how to better process these samples to optimize the sensitivity when detecting disease still exist. This study aimed to describe the impact of different tongue tips processing and testing protocols to optimize the sensitivity and specificity of PRRSV detection in sow herds. Samples from seven farms were tested using different pooling strategies, processing techniques, and storage conditions. Results showed that testing tongue tip fluids yielded more sensitive PRRSV detection compared to tongue tissue homogenates, and that keeping samples frozen yield lower Ct values when compared to refrigerated samples. Pooling samples reduced diagnostic accuracy but can still provide valuable information depending on the question being addressed. We suggest that practitioners discuss testing objectives with pathologists before sample submission. We also estimate that for every day elapsed since tongue tip collection, the Ct value increase by 0.2 units, suggesting that delays in sending the samples and shipment should be avoided. Tongue tips are an easy-to-collect sample type that targets animals potentially more likely to be infected (dead piglets), diminishing welfare concerns during sample collection. This study provides valuable insights into how testing choices and submission circumstances impact RT-PCR PRRSV testing results of tongue tips.
Project #: 23-078 | Principal Investigator: Cesar A. Corzo | Institution: University of Minnesota | Posted: 3/13/2025 | Keywords: Monitoring, Swine Health, PRRS, PEDv, Senecavirus, SVA, PRRSv 1H.18, PEDv Stability
Objective 1: Monitor trends in pathogens incidence and prevalence – PRRSv, PEDv, PDCoV, Senecavirus and central nervous system associated viruses continued to be monitored, each maintaining historical patterns. During 2023-2024 we explored and developed a method to estimate the breeding herd Senecavirus cumulative incidence. Fortunately, cumulative incidence remained below 2.5%, with most of the years remaining below 0.5%. Another objective was to estimate the time herds required to eliminate PEDV and explored associated factors. A significant reduction in time to consistently wean RT-PCR negative piglet was observed when comparing epidemic (i.e., 24 weeks) versus endemic (i.e., 13 weeks) stages of the disease in the US. Factors such as previous immunity, herd size, season when outbreak occurred were associated with the time to wean negative PCR piglets.
Objective 2: To conduct prospective monitoring of PRRSv sequence evolution and impact – During 2023-2024 we continued to curate our PRRSv ORF5 database. The representativeness of this database has enabled multiple collaborations, including outbreak investigations and, most recently, the development of the new PRRSv classification system. Thanks to this new classification we were able to identify a new variant of concern allowing us to communicate to the industry of this finding in a timely manner. Fortunately, this variant does not seem to have the transmissibility that the L1C.5 had; however, vigilance remains necessary. Lastly, we developed a mechanism to identify herds that are having a prolonged time-to-stability and are currently beta testing this methodology.
Objective 3: To expand participation of producers to allow for all to be involved – During 2023-2024 we added one production system and are awaiting enrollment forms from a second system. In addition, our website continued to be finetuned and updated with the information our readers are requesting, and we have reached over 15,000 views. The most visited section is related to reports, totaling 1,400 views. Interestingly, we are seeing that MSHMP website visitors are located around the world, but both the US and China are the main consumers. During 2024, our team and collaborators published a total of nine peer reviewed manuscript which speaks for the relevance and versatility of the dataset.
Project #: 23-029 | Principal Investigator: Gustavo Silva | Institution: Iowa State University | Posted: 3/11/2025 | Keywords: Biosecurity, wean-to-harvest, PRRSV, PEDV, swine
Objectives: The main objectives of this proposal were: 1) to assess the current bioexclusion practices used at wean-to-harvest sites across the U.S., ensuring a diverse group of producers from different swine-producing states are included; and 2) develop a tool that veterinarians, production managers, and producers can use to assess biosecurity on their sites quickly.
Research methodology: This study assessed biosecurity practices at wean-to-harvest sites across major U.S. pork-producing states. Data were collected through a questionnaire completed by 21 herd veterinarians, including production systems and independent producers. The questionnaire was developed with input from industry experts and comprised 69 questions on bioexclusion practices, covering site characteristics, vehicle movements, people movement, manure removal, water entry, and sanitation. A weighted method ensured the results reflected all respondents’ answers. For the second phase, we enrolled 139 wean-to-harvest sites to assess biosecurity practices and their relationship with lateral disease introduction of PRRS, PEDV, PDCoV, and TGEV. Farms must be stable or negative for PRRS and key enteric viruses, including PEDV, PDCoV, and TGEV. Participating producers were asked to complete a biosecurity questionnaire with 115 questions covering risk events, biosecurity management practices, herd demographics, trucking sanitation, and farm location.
Research findings: the results of phase 1 include data from 15.7 million pigs across 3,680 sites in 13 states. Of the 3,680 sites, 10.3% were nurseries, finishing represented 52.9%, and 36.8% were wean-to-finish sites. 93.3% of the farms reported using all-in-all-out, mortality disposal was mostly off-site (65.3%), and 47.3% of employees visited more than one site daily. While most sites have shower facilities (63.8%), fewer require employees to shower in (57.6%) or out (56.9%). Manure is removed about 1.5 times yearly, often by third-party companies. Most sites rely on well water (87.7%), but some don’t perform any water treatment (64.7%). Trucks hauling pigs are generally washed and disinfected, with 100% of trucks hauling weaned pigs cleaned between loads. For feeder trucks, 60.9% are washed, and 63.9% are disinfected between every load, and for market hog trucks, 78.3% are washed, and 52% are disinfected between every load. For the second phase, the data of 139 sites across nine companies in six states enhanced 44 nurseries from three companies, 44 finishers from three companies, and 51 grow-finish sites. The data shows that PRRSV outbreak rates were highest in grow-finish sites (61.4% – 27/44), followed by wean-to-finish (52.9% – 27/51) and nurseries (34.1% – 15/44). No outbreaks of PEDV or coronaviruses were reported in nurseries or wean-to-finish sites but grow-finish sites had a break rate for coronaviruses (2.3%) and a higher rate for PEDV (11.4%). Key findings include that nursery sites had 92% lower odds of reporting a PRRSV outbreak than finishers, and biosecurity practices like bench entry, truck washing, and downtime between loads reduced outbreak risk. Hauling animals with unknown status for PRRSV increased the odds of reporting an outbreak by 12 times, stressing the need for careful animal health monitoring before transportation.
Industry implications: The study’s first phase highlighted that while most sites reported implementing biosecurity measures like vehicle washing and employee training, gaps still need to be addressed, especially in communication and compliance auditing. The second phase revealed that nursery sites have a significantly lower risk of PRRSV outbreaks than grow-finish sites, which face a much higher risk. This emphasizes the need for stronger biosecurity in the finisher phase. Simple, cost-effective measures like bench entry—where employees change footwear or clothing before entering different areas—can help reduce the spread of PRRSV and are easy to implement. While these early findings are promising, more data is needed to refine biosecurity recommendations and help producers improve their practices, enhance surveillance, and build a more resilient industry.
Project #: 23-063 | Principal Investigator: Cesar A. Corzo | Institution: University of Minnesota | Posted: 2/19/25 | Keywords: Post-mortem, tongue tip, growing pigs, alternative specimens
In the US swine industry, most post-weaning health monitoring sampling relies on oral fluid sample collection since jugular venipuncture can be time-consuming and requires skilled personnel. Assessing whether easy-to-collect post-mortem samples, can provide value for diagnosis is needed as labor constrains are a concern in today’s industry. Furthermore, finding practical and time-efficient methodologies to monitor health during the post-weaning stages is necessary as this methodology can be rapidly adopted by the industry leading to a better understanding of disease dynamics. Recently, European researchers reported for the first time on the use of tongue tip fluids (TTF) sampling to detect PRRS given that their laws prohibit them from castrating piglets, thus, processing fluids was not an option for monitoring. In the US, work on TTF has been in preweaning animals but data on post weaning pigs is virtually non-existent. Therefore, the objectives of this study were 1) to assess the sensitivity and specificity of TTF, and other specimens including intracardiac blood (IC), oral/nasal swabs (ONS), rectal swabs (RS) and superficial inguinal lymph nodes (SILN) in growing pigs for the detection of PRRSV; and, 2) to characterize the detection of Porcine Circovirus type 2 and 3 (PCV2, PCV3), Porcine Parvovirus type 1 and 2 (PPV1, PPV2), Lawsonia intracellularis (Li), and Influenza A virus (IAV) in TTF, TTF, ONS, RS, and SILN.
Two growing pig farms located in Minnesota and representative of current swine production practices were included in this study. The first farm, a 2,400-head wean-to-finish farm undergoing a PRRS outbreak was visited when pigs were 6 and 12 weeks of age (WOA). The second farm was a 3,300-head finishing site undergoing a similar health challenge as farm 1 and was visited when pigs were 15 WOA. During each farm visit, a total of 30 dead pigs were sampled, resulting in a total sample of 90 pigs. From each pig, TTF, IC, ONS, RS and SILN samples were collected and tested in a way that for specific pathogens a gold standard sample was selected and then compare it with TTF results. Briefly, all TTF samples were tested individually by RT-PCR for all pathogens (i.e., PRRSV, SIV, PCV2/3, PPV1/2 and Li). All specimens were tested individually for PRRSV, while additionally ONS was individually tested for IAV; RS for PPV1, PPV2, and Li; while SILN were individually tested for PCV2 and PCV3. The sensitivity (Se), specificity (Sp), positive predictive value (PPV), and negative predictive value (NPV) were calculated for PRRSV TTF and IC serum as the gold standard. The proportion of RT-PCR positive results from specimens tested for other pathogens was compared by descriptive statistics. Most pathogens were detected at least once in TTF with Ct values ranging from 11.6 to 39.8. For PPRSV the best results for all specimens were at 11 WOA, when TTF had Se=84%, Sp=9%, PPV=62%, NPV=25%, ONS had Se=74%, Sp=73%, PPV=82%, NPV=62%, and SILN had Se=100%, Sp=9%, PPV=66%, NPV=100%. PCV2 was detected in 43% of TTF and 11% of SILN samples, PCV3 was not detected in any sample. PPV1 was detected in 1% of TTF and 0% of RS, PPV2 was detected in 97% of TTF and 61% of the RS samples. Li was detected in 6% of TTF and 0% of the RS samples. IAV was detected in 38% of TTF and 38% of the ONS samples.
Most pathogens were detected on TTF samples during the three different ages indicating that this specimen can provide valuable post-mortem information during a diagnostic investigation. Detection of some pathogens in TTF could be the result of shedding or contamination which should encourage practitioners and veterinarians to interpret results with caution when using TTF. In our case, the overall diagnostic performance of all the other specimens used besides TTF still requires further investigation. Indeed, a complete and exhaustive collection of multiple clinical specimens from different body systems remains the standard in diagnostic investigations.
Contact: [email protected]
Project #: 23-052 | Principal Investigator: John J McGlone | Institution: Texas Tech University | Posted: 2/19/25 | Keywords: Pigs, Vaccination, Environmental Enrichment, Erysipelas, Ileitis
Environmental enrichment (EE) devices or programs are required in some countries and in some markets. Any EE device that has a second purpose would more likely encourage adoption. We developed an EE device that allows pigs to self-administer liquids by building the EE device consistent with pig rooting, investigating and play behaviors. A pilot study demonstrated pig preference for the EE device when a maternal pheromone was sprayed on the device. We previously have shown that this method of vaccine delivery was efficacious for pigs to self-deliver a Salmonella vaccine. In this study, we sought to determine if the EE self-administration device could deliver vaccines for four diseases common among growing pigs. A baseline sample determined the antibody status of subjects. Assay for serum IgG and IgA determined efficacy with three treatments groups: (1) a control that was not vaccinated, (2) a group in which pigs were individually vaccinated by oral gavage or intramuscular (i.m.) injection, and (3) a self-vaccinated group. Self-vaccination was efficacious for Erysipelas and Ileitis vaccines in that pigs built robust titers to these antigens. Self-vaccination using commercially licensed vaccines at labeled doses and timing for Influenza and Mycoplasma hyopneumoniae did not stimulate serum or oral fluid antibodies. Using EE for self-vaccination of selected vaccines is possible today which will reduce labor needs, eliminate the need for needles, and will allow pen-level vaccinations or delivery of other animal health products (drugs, pheromones, etc.).
Project #: 22-003 | Principal Investigator: Yi Lu | Institution: University of Texas at Austin | Posted: 2/14/25 | Keywords: aptamers, PRRSV, PEDV, sensors, detection
Swine viral pathogens, such as Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and Porcine Epidemic Diarrhea Virus (PEDV), pose significant economic challenges to the global swine industry. To ensure prompt and effective management of these swine viral outbreaks, novel diagnostic tools that allow on-site and real-time detection are required. While diagnostic methods have been developed and are available, they either require sample pretreatment and a skilled operator performed on a costly equipment that takes hours to days in a professional laboratory and thus not suitable for on-site detection, or they cannot tell whether the virus in infectious or not, causing delays in managing the viral outbreak.
To meet these challenges, we have developed a novel method for direct detection of intact viruses without any sample pretreatment, with the ability to detect and differentiate infectious swine viruses such as PRRSV and PEDV from the same virus that has been rendered noninfectious by disinfection. The method is based on DNA aptamers that can be selected to bind and differentiate infectious swine viruses from noninfectious and other viruses. By immobilizing the aptamers into a nanopore, only infectious PRRSV or PEDV will produce electrochemical signal changes and thus can be detected and quantified using a handheld meter.
Our primary objectives for this project are as follows:
1. To obtain DNA aptamers that can bind infectious PRRSV and PEDV through in vitro selection and counter selection processes, with the aim of enhancing selectivity.
2. To design and validate DNA aptamer-nanopore sensors for the direct detection of infectious PRRSV and PEDV, both within controlled cell cultures and real-world field samples.
To achieve the objectives of this project, we conducted ten SELEX (Systematic Evolution of Ligands by Exponential Enrichment) experiments under varying conditions to optimize aptamer selection for porcine reproductive and respiratory syndrome virus (PRRSV). Through this process, we identified virus purity as critical to ensuring efficient aptamer selection throughout SELEX. Sequencing of SELEX DNA pools, followed by bioinformatic analysis, enabled us to identify promising aptamer candidates targeting PRRSV. Biochemical characterization of these candidates demonstrated that several could selectively bind to infectious PRRSV II, with minimal binding to noninfectious PRRSV II. While promising in vitro, additional optimization or complementary techniques may be required for field applications. The insights gained from this study underscore the importance of sample purity and the need to further enhance aptamer selectivity to improve detection reliability in applied settings.
Contact info for PI of the project: [email protected]
Project #: 22-059 | Principal Investigator: Gustavo Machado | Institution: North Carolina State University | Posted: 1/1/25 | Keywords: Truck, transport, disease modeling, contact trace, indirect contact, truck cleaning, and disinfection.
Jason A. Galvis1 and Gustavo Machado1
1Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, USA.
Introduction: Disease transmission via farm-to-farm transportation vehicles is still to be fully appreciated as a significant route of disease dissemination; however, it has slowly sounded the alarm for swine and systems. While transmission by vehicle movements has been associated with outbreaks such as foot and mouth disease (FMD) and African swine fever (ASF), it is still unknown how these movements represent a risk for disease transmission. This is due to the absence of information about pathogen viability on vehicle surfaces and the efficacy of cleaning and disinfection to eliminate such pathogens.
Methodology: In this study, we developed a novel methodology to reproduce the indirect contact network among farms by vehicle movements, which combines pathogen stability, conditioned to decay over time, with environmental temperatures. In addition, we included that pathogens may be eliminated by an effective cleaning and disinfection event. Because we did not know the actual efficacy of cleaning and disinfection procedures, we simulated six different cleaning efficacies: 0, 10%, 50%, 80%, 90%, and 100%, which are triggered each time a vehicle contacts a cleaning station. To ensure vehicles are a source of infection due to their proximity to the farms, we identified farm locations using the Perimeter Buffer Area (PBA) collected from RABappTM. Thus, a vehicle contacted a farm if it came within a determined distance from the PBA. To test this methodology, we collected movements from 567 vehicles across three swine companies in two regions of the U.S. These vehicles were divided into six transportation types depending on if they transported people, pigs to farms, pigs to market, feed, undefined, and one additional type combining all vehicles. For each vehicle type, we reconstructed a contact network and evaluated the median number of farms that could be infected by considering the chronological order of contact for one year.
Results: In region one, after a year of movements and without effective cleaning (0%), the combined vehicles were able to potentially infect a maximum of 2,157 farms, similar to the results of vehicles transporting feed (Figure 1). This result was lower for vehicles transporting pigs to farms (2,089), pigs to market (1,507), undefined vehicles (1,760), and personnel (3). However, with 100% cleaning efficacy, the farms in the contact chain of pooled vehicles, vehicles transporting feed, and undefined vehicles was reduced by 1%. This reduction continued with 26% for vehicles transporting pigs to farms, 43% for vehicles transporting pigs to market, and 66% for vehicles transporting crew. In region two, which only had one vehicle transportation role with an inefficient cleaning efficacy (0%), undefined vehicles can potentially infect 437 farms, and this number decreased by 76% with 100% cleaning efficacy.
Discussion and conclusion: Except for vehicles transporting crew, all other vehicle types exhibited the potential to disseminate disease to numerous farms in both regions. For vehicles
transporting feed and undefined vehicles in region one, increased cleaning efficacy showed a low impact in reducing potentially infected farms, which is associated with a low frequency of cleaning events. On the contrary, for vehicles transporting pigs to farms, pigs to market, and undefined vehicles in region two, an increased cleaning efficacy showed a higher reduction of potentially infected farms due to a higher frequency of which trucks drove through cleaning stations. This is likely related to a higher perception of risk due to these vehicles having direct contact with organic material by transporting live animals. In conclusion, even when vehicle cleaning and disinfection is fixed at 100% efficacy, vehicles can create numerous paths in the network to spread swine diseases. Thus, new strategies are necessary to reduce the risk of transmission by vehicle movement, such as redirecting routes or increasing the frequency of cleaning.
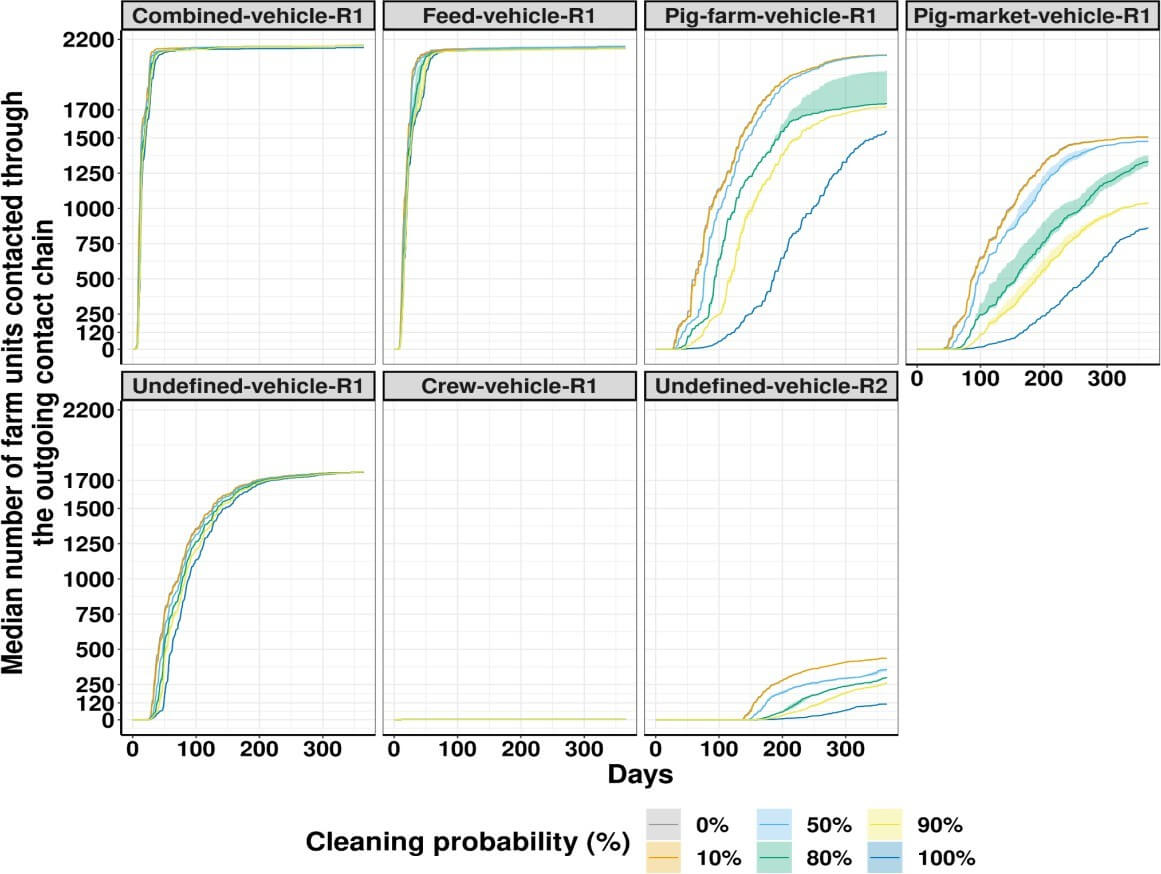
Figure 1. The number of farm units potentially infected via vehicle movements. Solid lines represent the median, while shadowed areas represent the interquartile ranges from ten simulations performed for each cleaning probability. Region one is identified by R1 and region two by R2. Combined-vehicles represent the combination of all vehicles movements from region one, feed- vehicle are vehicles transporting feed to farms, pig-farm-vehicle are vehicles transporting live pigs to farms, pig-market-vehicle are vehicles transporting live pigs to market, crew-vehicle vehicles are vehicles transporting farm personnel, and undefined-vehicles are vehicles without a defined role.
Project #: 23-076 | Principal Investigators: Isadora Machado, DVM MSc., and Daniel Linhares, DVM MBA PhD. | Institution: Iowa State University | Posted: 12/13/24 | Keywords: PRRSV, tongue fluids, risk-based, monitoring, targeted sampling, swine
Objectives: The primary objective of this study was to evaluate tongue fluids (TF) as a risk-based sampling approach in commercial breeding herds. TF PRRSV reverse transcription-qualitative polymerase chain reaction (RT-qPCR) results from stillborn piglets were evaluated as an indicator of PRRSV RNA detection in liveborn littermates. Secondly, we evaluated the presence of stillborns and litter size as indicators of PRRSV detection in live piglets.
Methodology: Samples were collected from two PRRSV-positive breeding herds (Herd A: 2,500-sow farm with a three-week batch farrowing system; Herd B: 4,500-sow farm with a weekly batch farrowing system). A total of 130 litters were sampled within 12 hours after farrowing: 66 without stillborn piglets and 64 with stillborn piglets, totaling 1,723 liveborn and 105 stillborn sampled piglets. Regarding the litter’s parity: in Herd A, 14 were parity 1 sows, 6 were parity 2, 4 were parity 3, and 1 parity 4; in Herd B, all 104 litters came from parity 1 sows.
TF and intracardiac blood (IB) samples were individually collected from stillborn piglets, and tail blood swabs (BS) were individually collected from all liveborn littermates within the selected litters. Samples were individually tested for PRRSV RNA detection by RT-qPCR at the Iowa State University Veterinary Diagnostic Laboratory. Litters with ≤ 11 liveborn piglets were defined as small litters, whereas litters with ≥ 12 were defined as large. Generalized linear regression models were used to evaluate:
– Incidence of stillborn piglets on the probability of PRRSV RNA detection within the litter;
– Stillborn TF PCR-result as an indicator of PRRSV in liveborn piglets;
– Litter size as a risk factor for PRRSV.
Research Findings: Considering all sample types (TF, IB, and/or BS), the percentage of PRRSV-positive litters based on PRRSV RNA testing was 21.5% (28 of 130 litters), of which 15.4% (20 of 130 litters) were BS PRRSV RNA-positive, 10% (13 of 130 litters) were IB PRRSV RNA-positive, and 16.9% (22 of 130 litters) were TF PRRSV RNA-positive. At an animal-level, considering both herds, 3.4% (59 of 1,723 liveborn piglets) were BS PRRSV-RNA positive, 16.2% (17 of 105 stillborn piglets) were IB PRRSV-RNA positive, and 26.7% (28 of 105 stillborn piglets) were TF PRRSV-RNA positive (Table 1). The mean positivity of liveborn piglets within the litter was 5%, varying from 0% to 85.7%, while the total born (stillborn and liveborn piglets combined) was 4.6% (0% to 76.4%).
Regarding sample types’ Ct values, BS had a median of 35.5 (range from 19.3 to 39.9), IB a mean of 22.4 (range from 16.1 to 36.7), and TF a mean of 29.2 (range from 24.6 to 38.5) (Figure 1). Intracardiac blood exhibited the lowest median Ct value, which was significantly different from the mean Ct values of the sample types (P-value < 0.05).
In litters with at least one stillborn compared to those without a stillborn, the odds of having a PRRSV RNA-positive result (IB, TF, or BS) increased 5.6 times (95% confidence interval [CI]: 2.21–16.28, P-value < 0.001), and the odds of having at least one viremic liveborn littermate (BS) increased 2.8 times (95% CI: 1.04–8.40, P-value = 0.049). The probability of detecting PRRSV in litters with stillborn was 35.9% (25.2%, 48.3%), compared to 9.1% (4.1%, 18.8%) in large litters. When the stillborn TF was positive, the odds of having a viremic liveborn littermate (BS) increased 20.8 times (95% CI: 6.90–69.56, P-value < 0.001).
Small litters had 5.3 times higher odds (95% CI: 2.21–13.13, P-value = 0.002) of yielding a positive result (IB, TF, or BS) than large litters. The probability of detecting PRRSV in small litters was 45.7%, compared to 13.7% in large litters. In small litters, the odds were 7.4 times higher (95 % CI: 2.72–21.91, P-value < 0.001) for having at least one positive liveborn piglet and 2.7 times higher (95 % CI: 1.05–7.19, P-value = 0.03) for having at least one positive stillborn piglet. Results are summarized in Table 2. Lastly, litter size using total born per litter had no confounding effect and was not statistically significant; therefore, it was not included in the final model. Results are summarized in Table 2.
Industry Implications: Litters with stillborn piglets and small litter size had a higher probability of being PRRSV RNA-positive. Moreover, TF samples from stillborn piglets are a reliable indicator of the PRRSV status of their liveborn littermates, indicating that TF can be an effective risk-based approach for PRRSV detection. Therefore, veterinarians and pig producers are encouraged to collect TF, targeting stillborn piglets and small litters to increase the likelihood of detecting PRRSV. This risk-based sampling strategy within the first hours post-farrowing enhances the effectiveness of PRRSV monitoring programs in breeding herds that support timely interventions, such as McREBEL and cross-fostering. Lastly, while TFs were individually collected in this study to test the hypothesis, collecting them as an aggregated sample, similar to processing fluids, is recommended. This approach increases the likelihood of detecting the virus, allowing for a larger number of animals to be screened.
Project #: 23-053 | Principal Investigator: Nubia Resende de Macedo | Institution: Iowa State University | Posted: 12/12/24 | Keywords: Glaesserella australis, Diagnostics, Swine, Novel, Pathogen
Industry Summary: Glaesserella australis, a newly recognized Gram-negative bacterium species, was isolated from the lung lesions of pigs in Australia in 2018. More recently, in 2023, G. australis was also identified in two swine herds in Ontario, Canada. The isolates were obtained from the pericardium and lung tissues of pigs. The hypothesis remains that G. australis has been misdiagnosed over the years since the disease presentation is similar to porcine pleuropneumonia caused by Actinobacillus pleuropneumoniae, and no methods were being used to detect G. australis accurately.
Sequencing of historical isolates with similar characteristics was attempted at the ISU VDL, but no match was found. Therefore, the G. australis reference strain was obtained and added to our sequencing platform and, most importantly, to our MALDI-TOF database for real-time screening of clinical samples. Additionally, an RT-PCR is now available for further screening and diagnostics purposes. An ISH assay is also being developed to detect this organism in tissues associated with lesions.
At the ISU VDL, since January of 2024, we have screened for G. australis through MALDI-TOF, but this new potential swine pathogen is yet to be found. Nevertheless, the tools described here will contribute to the G. australis screening process in the U.S. and help improve our understanding of its prevalence and pathogenesis.
Project #: 23-028 | Principal Investigator: Gustavo Machado | Institution: North Carolina State University | Posted: 1/10/25 | Keywords: Trucks, deliveries, transportation, prevention strategies, and biosecurity
Jason A. Galvis1, Cesar Corzo2 and Gustavo Machado1
1Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, USA.
2Veterinary Population Medicine Department, College of Veterinary Medicine, University of Minnesota, St Paul, MN, USA
Introduction: Substantial evidence indicates that vehicle movement is closely linked to the spread of diseases among animal production sites. To mitigate these transmission events, vehicles undergo thorough cleaning and disinfection (C&D) procedures. However, C&D effectiveness remains an open-ended question, while the frequency of C&D between farm visits is unknown. Consequently, relying solely on vehicle C&D is insufficient to control the spread of diseases, and supplementary strategies are necessary to prevent disease transmission events via contaminated vehicles. The objective of this study was to reduce the risk of between-farm transmission through vehicle contacts by rerouting vehicles while considering C&D events and effectiveness.
Methodology: We collected movements of 654 vehicles in a pig-dense area in the U.S. and identified vehicles visiting farms, C&D, slaughterhouses, feed mills, and parking locations. Farm data was collected from enhanced on-farm Secure Pork Supply (SPS) biosecurity plans available in the Rapid Access Biosecurity application (RABappTM). We ranked and reorganized the vehicles delivering animals and feed to farms according to several conditions, including disease status of visited farms, vehicle contact network communities, C&D events, and shipment time efficiency. Using these conditions, we simulated vehicle movements for one week, indicating each vehicle was C&D after each shipment. We reconstructed the between-farm contact network by vehicle movements from observed and simulated data and compared i) the number of contacts from PRRSV-positive and PEDV-positive farms to disease-free farms and ii) contacts between farms from different network communities (group of farms densely interconnected). In addition, we calculated the frequency of vehicles visiting C&D stations and traveled distances.
Results: Implementing our rerouting system led to a substantial decrease in the median number of risky contacts among farms (Figure 1). For vehicles transporting feed, risky contacts were reduced by 42% when C&D effectiveness was 0% and 89% when C&D was 50%. For vehicles transporting pigs to market, risky contacts were reduced by 25%, with C&D effectiveness at 0% and 45% with C&D at 50%. Vehicles transporting pigs between farms only showed a remarkable reduction after C&D effectiveness above 50%, with 33% fewer risky contacts. Finally, with C&D effectiveness at 100%, risky contacts dropped below 5% for vehicles transporting feed and pigs to market, and below 37% for vehicles transporting pigs to farms. Our system also reduced the interactions between farms from distinct network communities, 17% when C&D effectiveness was 0%, and 99.9% with C&D effectiveness at 100%. Furthermore, the rerouting system increased by up to 81% in C&D visits and up to 54% in distance traveled per vehicle.
Discussion and conclusion: We showed that by rerouting vehicle movements, risky contacts among farms were reduced. Despite the potential benefit of preventing disease spread, our rerouting system increased the C&D events and the distance traveled per vehicle. Given the severe economic impact of PRRSV, PEDV, ASFV, and other endemic infectious diseases on swine production, the costs and logistics of a vehicle rerouting system require a close examination to justify the potential economic benefits of reducing disease transmission compared to continuing traditional vehicle movement schedules and C&D protocols. Ultimately, our rerouting system could be integrated into regular vehicle shipment schedule operations as an additional tool for preventing and controlling the spread of livestock diseases among farms via the indirect contact network of vehicle movements
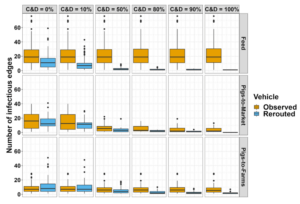
Figure 1. Infectious edges within the vehicle contact network. Box plots displaying the number of connections, named infectious edges or risky contact, from PRRSV-positive and PEDV-positive farms to disease-free farms within the vehicle contact network for one week.
Project #: 23-036 | Principal Investigator: Rodger Main, DVM, PhD | Institution: Iowa State University | Posted: 1/10/25 | Keywords: PRRSV, PEDV, Biosecurity, Temperature, Time.
Since its introduction in 2013, PEDV remains an important disease to the US swine industry, necessitating robust biosecurity measures. This study aimed to evaluate various cleaning protocols—Positive Control (no cleaning), Dry Clean – Scrape and Bake (TADD), Volume Hose Wash and Disinfect, Power Wash and Disinfect, and Negative Control—to determine their efficacy in mitigating PEDV spread from a contaminated trailer to the farm site area after simulating foot traffic between these two areas.
Livestock trailers were contaminated with PEDV-positive feces, except for the Negative Control using PEDV-negative feces. Different cleaning methods were applied, followed by a simulation to mimic real-world conditions. PEDV presence was quantified using qPCR to measure viral load on trailer surfaces and farm loading areas. Bioassay was conducted to inoculated naïve pigs with samples recovered from the farm site area after applying the treatments on the trailers. Statistical analyses, including ANOVA and Fisher’s Exact test, compared PEDV levels and positive PCR results across treatments.
Washing treatments, particularly volume hose and power wash & disinfect, demonstrated significant efficacy in reducing viral load on both trailer and farm-site surfaces. On farm-site surfaces, these methods achieved over 99% reduction in viral genomic copies compared to the positive control. This marked reduction is crucial in preventing infection of susceptible pigs and highlights the importance of effective washing protocols. The Power Wash and Disinfect method emerged as highly effective, significantly reducing PEDV levels on trailer surfaces with most samples showing negative PCR results (3 of 5). The Volume Hose Wash and Disinfect method also demonstrated substantial efficacy in inactivating the virus on the bioassay, though some residual fecal material was observed. Even though the Scrape and Bake method reduced the viral load in more than (>98%), it was not effective in terms of virus inactivation, where the pigs from four out of five replicates became infected for PEDV on bioassay. All negative control replicates were negative in the qPCR result of the surfaces sampled and remained negative on the bioassay after inoculation.
The findings from this study suggest routinely cleaning and disinfecting all market haul trailers leaving terminal points of concentration by either of the water-based trailer cleaning treatments evaluated present as an opportunity to create a step-change in the magnitude of inter-premises disease transmission associated with market haul transport and elevate the state of preparedness across the US pork industry.
Contact: [email protected]
Project #: 23-070 | Principal Investigator: Gustavo De Sousa E Silva | Institution: Iowa State University | Posted: 1/10/25 | Keywords: PRRSV, perinatal, mortalities, tongue, virus isolation, swine
Objectives: This study aimed to: 1) Compare the detection of the Porcine reproductive and respiratory syndrome virus (PRRSV) and Influenza A virus (IAV) using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in weekly oral fluids (OF) and TF from wean-to-market pigs, 2) Assess the likelihood of successful PRRSV ORF-5 sequencing from these two sample types (OF vs. TF), 3) Assess the effect of pooling TF on the RT-qPCR detection of PRRSV.
Research Methodology: This study monitored three groups of pigs from two production systems, testing for PRRSV and influenza A virus (IAV) from weaning until market. Two groups (A and B) were sourced from PRRSV-positive stable sow farms, while Group C was from a farm weaning positive pig. Groups B and C received a PRRSV-modified live virus (MLV) vaccine at weaning. Weekly, oral fluid (OF) samples from six pens and TF samples were tested for PRRSV and IAV using RT-qPCR. PRRSV-positive TF and OF samples were selected and Sanger sequenced to compare the performance of both sample types. PRRSV-positive TF samples were also serially diluted to assess the impact of pooling on PRRSV detection. The study evaluated associations between weekly pathogen detection and mortality using mixed-effects regression models, as well as diagnostic accuracy and agreement between OF and TF results. Additionally, statistical models were used to assess the cycle threshold (Ct) changes with varying dilution factors to determine the minimum dilution level at which PRRSV detection was maintained in 95% of cases.
Research Findings: There were 60 weeks across all three groups with sample submissions, OF samples were obtained in all 60 weeks, and TF samples were collected in 43 of those weeks. IAV was detected in 34.9% of OF and 30.2% of TF samples, while PRRSV was found in 67.4% of OF and 53.5% of TF samples. TF samples showed a significantly lower mean cycle threshold (Ct) for PRRSV (28.7) compared to OF samples (33.2). There was fair agreement between the RT-qPCR outcomes of TF and OF (Cohen’s kappa values of 0.26 for PRRSV and 0.24 for IAV). Out of the 22-week-matched TF and OF pairs sent for PRRSV sequencing, 45.5% of OF and 63.6% of TF samples were successfully sequenced. The higher success of sequencing in TF samples was attributed to their relatively lower Cts. For both sample types, mortality rates were significantly higher when PRRSV was detected, especially when detected alongside IAV. Additionally, dilution studies demonstrated that pooling samples could effectively increase detection probabilities, with 79% detection at a 1/7 dilution compared to 14.29% detection probability if only one day’s TF out of one week is tested for the same scenario of 1 positive day.
Industry Implications: Tongue fluid (TF) samples offer a comparable and cost-effective alternative to oral fluid (OF) samples for the RT-qPCR detection of PRRS virus and Influenza A Virus (IAV) in growing pig herds, especially when dead animals are available. Despite PRRSV and IAV RT-qPCR detection being numerically more frequent in OF, TF samples had a higher success rate for PRRSV sequencing due to their significantly lower Ct values. This makes TF a useful surveillance sample type for PRRSV in wean-to-finish barns. To maximize the diagnostic potential of this sampling approach, practitioners are encouraged to aggregate tongue tissues from all daily mortalities. As there could be weeks without mortality, postmortem TF sampling and antemortem sampling (e.g., OF) can be jointly part of a surveillance program. Furthermore, testing TF pools in weekly pools can be useful in wean-to-finish barns where budget constraints may limit testing options.
Project #: 23-051 | Principal Investigators: James F. Lowe | Institution: Lowe Consulting Ltd. | Posted: 12/16/24 | Keywords: Swine biosecurity, Trailer decontamination, Disease transmission, Epidemiological modeling, Cost-benefit analysis
Objectives: The primary goal of this study was to determine the most effective and cost-efficient way to clean market haul trailers that transport pigs between wean-to-finish farms and slaughter facilities. We aimed to understand how different levels of trailer washing impact the spread of Porcine Epidemic Diarrhea virus (PEDv) and to find the best practices that balance disease control and economic feasibility.
Research Conducted: We used computer simulations to model swine production systems under various conditions. The first scenario focused on a single production system with 24,000 sows across 8 sites. The second scenario looked at a region with 24,000 sows divided into 4 systems, each geographically related but otherwise unconnected. We ran simulations to see how washing different percentages of trailers would affect the spread of PEDv. Additionally, we tested scenarios with different PEDv prevalence levels to see how these conditions would change the effectiveness of trailer washing.
Findings: 1. Single Production System:
– Washing 100% of trailers significantly reduced the number of infected farms, with an average of 23.13 infected premises.
– Washing 60% of trailers was identified as a cost-effective strategy, achieving a significant reduction in disease spread at a lower cost of about $32,956 per farm.
2. Geographically Related Systems:
– Washing 100% of trailers in this setup reduced the average number of infected premises to 10.56.
– Surprisingly, washing 0% of trailers was found to be the most cost-effective in these systems, costing about $25,664 per farm, suggesting that extensive decontamination might not always be necessary in such segregated systems.
3. Impact of Disease Prevalence:
– In scenarios with high PEDv prevalence (20%), it was necessary to wash 80% of trailers to achieve a significant reduction in infection, at a cost of $48,957 per farm.
– In low prevalence scenarios (5%), washing trailers was not required to minimize costs, which dropped to $26,862 per farm.
What These Findings Mean for the Industry:
The study shows that cleaning market haul trailers is crucial for controlling the spread of PEDv, but the extent of washing needed can vary. For interconnected systems or during high prevalence periods, thorough washing of trailers is essential. However, in more isolated systems or when disease prevalence is low, producers might save costs without compromising biosecurity by washing fewer trailers. These insights can help producers develop tailored cleaning protocols that enhance swine health and productivity while managing costs effectively.
By adopting these findings, producers can make informed decisions about trailer decontamination practices, ensuring their operations are both economically viable and biosecure. This research provides a practical roadmap for reducing PEDv transmission and safeguarding the health of swine herds.
Project #: N/A | Principal Investigators: Montse Torremorell, Juan Mena, Ana Marcos, Marie Culhane | Institution: University of Minnesota | Posted: 10/17/2024 | Keywords: HPAI, H5N1, influenza, literature review
The recent emergence of high pathogenic avian influenza (HPAI) H5N1 clade 2.3.4.4b in dairy and the ongoing outbreaks of HPAI in commercial poultry seriously threaten the U.S. swine industry. Due to the role that pigs play in the overall ecology of influenza infections, the potential risk of H5 infections in people, and the epidemiological links between swine, dairy, and poultry, it is of the utmost importance to fully understand the risks to pigs and from pigs to other species, including humans. Of equal importance is the development of science-based strategies needed to prevent the introduction of H5N1 into pigs and contain it should incursions occur.
Project #: 23-067 SHIC | Principal Investigator: Onyekachukwu Henry Osemeke | Institution: Iowa State University | Posted: 9/27/2024 | Keywords: PRRSV, perinatal, mortalities, tongue, virus isolation, swine
Objectives: This study aimed to assess different sample collection protocols and cell lines for successfully isolating live porcine reproductive and respiratory syndrome virus (PRRSV) from perinatal piglet mortalities (stillborn piglets and piglets that died within 24 hours of birth) using tongue tissue fluids (TF).
Research Methodology: Samples were obtained from 597 perinatal mortalities in a 5,000-head PRRSV-positive breeding herd over a 4-day period. Tongue tissues were grouped into 20 batches (approximately 30 mortalities or tongues per batch). Each tongue was divided into four quarters, with each quarter randomly assigned to one of four collection protocols: 1) TF extraction from fresh tissues using Phosphate Buffered Saline (PBS), 2) TF extraction from fresh tissues using Virus Transportation Medium (VTM), 3) TF extraction in PBS after one freeze-thaw cycle (Freeze-thaw), and 4) using tongue tissue homogenate (Homogenate). This resulted in 80 samples total (20 batches x 4 protocols), all sent to a NAHLN-approved veterinary diagnostic laboratory for RT-qPCR testing. The RT-qPCR cycle threshold (Ct) values were averaged across the four protocols in each of the 20 batches, and the 10 batches with the lowest mean Ct values were selected for virus isolation (VI). Two cell lines (ZMAC and MARC-145) and one batch of primary alveolar macrophages (PAM) were tested for their effect on successful PRRSV isolation.
Research Findings: All samples tested positive for PRRSV by RT-qPCR; the average Ct values for the PBS, VTM, Freeze-thaw, and Homogenate groups were 21.9, 21.8, 22.6, and 24.8, respectively.
PRRSV was isolated successfully from tongue tissues in all groups with varying success rates. The virus isolation success rate was 22.6% in the PBS group, 12.1% in the VTM group, and 2.8% in both the Freeze-thaw and Homogenate groups. The probability of successful viral isolation was 3.1% in MARC-145 cells, 21.0% in ZMAC cells, and 4.8% in PAM cells. Mortality batches with only stillborn piglets had a 35.5% probability of successful PRRSV isolation, while batches with stillborn and dead piglets had a 1.0%.
Industry Implications: Live PRRSV can be isolated from postmortem tongue fluids. Extracting TF from fresh stillborn piglets using PBS or VTM increases the chances of successful virus isolation. The ZMAC cell line outperformed the MARC-145 cell line and PAM cells in this study. Ensuring a cold chain from sample collection until arrival at the laboratory maintains the diagnostic quality of the samples. Isolating PRRSV from aggregate samples such as TF provides several surveillance and vaccine development benefits. Virus isolation using aggregate samples allows for the efficient co-detection of multiple PRRSV strains within a herd if present, this has great advantages for surveillance and developing autogenous vaccines.
Project #: 23-079 SHIC | Principal Investigator: Mariana Kikuti, Cesar A. Corzo | Institution: University of Minnesota | Posted: 9/27/2024 | Keywords: postmortem sampling; PRRSV detection; specimen sensitivity; disease monitoring; diagnostic accuracy
Specimens collected from dead pigs are a welfare-friendly and cost-effective active surveillance. This study aimed to evaluate the accuracy of different postmortem specimens from dead piglets for disease detection, using PRRSV as an example. Three farrow-to-wean farms undergoing PRRSV elimination were conveniently selected. Samples were collected at approximately 8- and 20-weeks post-outbreak. Postmortem specimens included nasal (NS), oral (OS), and rectal (RS) swabs, tongue-tip fluids (TTF), superficial inguinal lymph nodes (SIL), and intracardiac blood. These were tested individually for PRRSV by RT-PCR. Sensitivity, specificity, negative and positive predictive values, and agreement of postmortem specimens were calculated using intracardiac sera as the gold standard. OS and SIL had the best overall performance, with sensitivities of 94.6–100%, specificities of 83.9–85.1%, and negative predictive values of 97.3–100%. TTF had high sensitivity (92.2%) but low specificity (53.9%) and positive predictive value (48.3%). While challenges in meeting sampling
targets due to variable pre-weaning mortality were noted, PRRS was detected in all postmortem specimens. OS and NS showed promising results for disease monitoring, though TTF, despite their sensitivity, had lower specificity, making them less suitable for individual infection assessment but useful for assessing environmental contamination.
Project #: 23-044 | Principal Investigator: Dr. Derald Holtkamp | Institution: Iowa State University | Posted: 8/20/2024 | Keywords: Standardized Outbreak Investigation Program, emerging disease, transboundary disease, Rapid Response Rrogram, epidemiology
Industry Summary: To protect the United States swine industry from substantial economic losses caused by emerging, or transboundary disease outbreaks, producers and veterinarians need to rapidly identify, control, and slow the spread of the pathogen. In response to events following the introduction of the porcine epidemic diarrhea virus (PEDV) into the United States in 2013, the Swine Health Information Center (SHIC) funded Iowa State University to develop the Rapid Response to Emerging Disease Program (RRP) in August 2016. The program now includes a nationwide network of veterinarians, state animal health officials or representatives, epidemiologists, and, when appropriate, federal animal health officials called the Rapid Response Team (RRT). RRT members are trained, prepared, and committed to moving within 24 hours of contact to conduct epidemiological investigations when a new transboundary or emerging disease threat occurs. The approach, methodology, form and reports were developed to systematically, consistently, and comprehensively conduct outbreak investigations for the RRP.
In 2021, SHIC funded the development of the Standardized Outbreak Investigation Program for the RRP (21-080 SHIC, 22-076 SHIC, 23). The form developed for the Standardized Outbreak Investigation Program to conduct outbreak investigations, formatted as a Word® (Microsoft, Redmond Washington) document, is available for download on the SHIC website at https://www.swinehealth.org/investigation-instrument/ (accessed May 29, 2024). A web-based application was developed for the Standardized Outbreak Investigation Program. The application was launched in January of 2024 and is available to RRT members and other veterinarians and producers to conduct outbreak investigations on sow farms.
The objective of this project was to provide ongoing management and coordination of the RRP to maintain the readiness of the RRT.
The transitional Project Coordinator, who was appointed in 2021, provided support to RRT members, assisted in conducting outbreak investigations of endemic diseases, and was available in the event of an animal health emergency where the RRT was called upon.
The principal investigator (Dr. Holtkamp), Transitional Project Coordinator (Chris Mowrer) and Dr. Kate Dion collaborated with SES Inc., who was contracted by SHIC, to develop nine training videos. to develop training for the RRT and other users of the Standardized Outbreak Investigation Program web application. The final version of these videos is expected by June of 2024.
Contact Derald Holtkamp at [email protected]
Scientific Abstract: Same as Industry Summary.
Introduction: To protect the United States swine industry from substantial economic losses caused by emerging, or transboundary disease outbreaks, producers and veterinarians need to rapidly identify, control, and slow the spread of the pathogen. In response to events following the introduction of the porcine epidemic diarrhea virus (PEDV) into the United States in 2013, the Swine Health Information Center (SHIC) funded Iowa State University to develop the Rapid Response to Emerging Disease Program (RRP) in August 2016. The program now includes a nationwide network of veterinarians, state animal health officials or representatives, epidemiologists, and, when appropriate, federal animal health officials called the Rapid Response Team (RRT). RRT members are trained, prepared, and committed to moving within 24 hours of contact to conduct epidemiological investigations when a new transboundary or emerging disease threat occurs. The approach, methodology, form and reports were developed to systematically, consistently, and comprehensively conduct outbreak investigations for the RRP.
In 2021, SHIC funded the development of the Standardized Outbreak Investigation Program for the RRP (21-080 SHIC, 22-076 SHIC, 23). The form developed for the Standardized Outbreak Investigation Program to conduct outbreak investigations, formatted as a Word® (Microsoft, Redmond Washington) document, is available for download on the SHIC website at https://www.swinehealth.org/investigation-instrument/ (accessed May 29, 2024). A web-based application was developed for the Standardized Outbreak Investigation Program. The application was launched in January of 2024 and is available to RRT members and other veterinarians and producers to conduct outbreak investigations on sow farms.
Objectives: The objective of this project was to provide ongoing management and coordination of the RRP to maintain the readiness of the RRT.
Materials & Methods: Support a Transitional Project Coordinator and investigations conducted by RRT members. Funding for this project provided support for a transitional Project Coordinator who was previously appointed in 2021. When requested by producers, RRT members were invited to conduct investigations of outbreaks of endemic diseases. The transitional Project Coordinator gathered the information needed to prepare the outbreak investigation form and coordinated the investigations with all of the relevant parties (RRT member, herd veterinarian, farm manager, etc.). The investigations provided opportunities for RRT members to practice investigations of outbreaks of endemic diseases.
Training of the RRT on the web-based industry-standard outbreak investigation instrument. SHIC contracted with SES Inc. to develop training for the RRT and other users of the Standardized Outbreak Investigation Program web application. The principal investigator (Dr. Holtkamp), Trasitional Project Coordinator (Chris Mowrer) and Dr. Kate Dion collaborated with SES to develop nine videos.
Results: Transitional Project Coordinator and investigations conducted by RRT members. The transitional Project Coordinator provided support to RRT members and assisted in conducting outbreak investigations of endemic diseases and was available in the event of an animal health emergency where the RRT we be called upon to conduct outbreak investigations. If such an event were to occur, it is expected that a full-time project coordinator would be required, as the number of investigations would increase significantly to assure the investigations were conducted quickly. Requests for RRT members to conduct SHIC funded outbreak investigations have declined. Only one request was received in 2023/2024, down from 13 in 2022/2023.
Training of the RRT on the web-based industry-standard outbreak investigation instrument. The investigators collaborated with SES, who was contracted by SHIC, to develop nine training videos. The final version of these videos is expected by June of 2024.
Discussion: The RRP aims to build the infrastructure to respond to emerging or transboundary disease threats rapidly to slow the spread of the pathogen. The value of that infrastructure depends on the RRT members’ ability to collect epidemiological information to identify hazards and characterize the geographic and temporal patterns of how the virus is spreading from one herd to another. Maintaining the readiness of the RRT is critical to realizing the value if an emerging or transboundary disease event occurs in the US.
Project #: SHIC 23-019 | Principal Investigators: Erin Kettelkamp, DVM | Institution: Swine Vet Center | Posted: 6/14/2024 | Keywords: aerosol, biocontainment, biosecurity, fan covering, PRRS, swine
Objective: The primary objective of this study was to evaluate the effectiveness of various rapidly deployable, exhaust fan cover materials in reducing airborne particles that can carry swine respiratory pathogens. These pathogens include viruses such as PRRSv (Porcine Reproductive and Respiratory Syndrome virus), PEDv (Porcine Epidemic Diarrhea virus), and IAV (Influenza A virus). By identifying effective and rapidly deployable fan coverings, we aim to enhance biosecurity measures and reduce the spread of these diseases within and between swine farms.
Research Conducted: The study was conducted at a commercial swine facility, focusing on three types of fan covers: PolyKlean™ synthetic air filter media (Blue Poly), a nylon tear-resistant fan sock (Fan Sock), and a polyethylene privacy screen material (Black Screen). These were compared to fans with no coverings (control). Airborne particle counts were measured using a handheld particle sizer to determine the reduction in particles ranging from 0.3 to 5.0 micrometers (µm). Measurements were taken at various distances from the fans to assess the effectiveness of each covering material. Weather conditions and ventilation settings were carefully monitored and recorded to ensure consistency across all sampling points.
Research Findings: The results showed that the Fan Sock was the most effective in reducing airborne particle quantities (0.7 to 5.0 µm) at 1 meter from the fan compared to the Blue Poly and control treatments, with the Black Screen treatment being intermediate. However, as the distance from the fan increased, differences in particle quantities were not observed across the treatments, resulting in no overall differences.
Implications for the Industry: These findings suggest that implementing exhaust fan coverings can be most beneficial at reducing larger air particles up to short distances (up to 1 meter) from fans. Based on the relationship between air particle size and the spread of airborne swine pathogens, additional research is warranted to understand the role of fan coverings on biocontainment. The fan sock provided better airflow and is already commonly used in the swine industry, making it a more practical option for rapid deployment during disease outbreaks to potentially improve regional biocontainment. Further research is recommended to validate these findings and explore additional biosecurity measures.
Project #: 24-015 | Principal Investigators: Pablo Piñeyro | Institution: Iowa State University | Posted: 7/9/2024 | Keywords: PCV4, In situ hybridization, Lymphoid tissue, Coinfection, PCV2, PCV3
Since PCV4 was first described in 2019, the virus has been identified in several countries in Southeast Asia and Europe. Most studies have been limited to detecting PCV4 by PCR. Thus, PCV4 has an unclear association with clinical disease. This study utilized 512 porcine clinical lung, feces, spleen, serum, lymphoid tissue, and fetus samples submitted to the ISU‑VDL from June–September 2023. PCV4 was detected in 8.6% of samples with an average Ct value of 33. While detection rates among sample types were variable, lymphoid tissue had the highest detection rate (18.7%). Two ORF2 sequences were obtained from lymphoid tissue samples and had 96.36–98.98% nucleotide identity with reference sequences. Direct detection of PCV4 by RNAscope revealed viral replication in B lymphocytes and macrophages in lymph node germinal centers and histiocytic and T lymphocyte infiltration in the lamina propria of the small intestine. PCV4 detection was most commonly observed in nursery to finishing aged pigs displaying respiratory and enteric disease. Coinfection with PCV2, PCV3, and other endemic pathogens was frequently observed, highlighting the complex interplay between different PCVs and their potential roles in disease pathogenesis. This study provides insights into the frequency of detection, tissue distribution, and genetic characteristics of PCV4 in the US.
Project #: 24-001 | Principal Investigator: Dustin Boler | Institution: Carthage Innovative Swine Solutions | Posted: 7/9/2024 | Keywords: ATP bioluminescence, disease monitoring, sow farm biosecurity, surveillance
This research evaluated the performance of three adenosine triphosphate (ATP) instruments to assess the cleanliness status of farrowing crates and the level of contamination after washing. Visual inspection to determine if a farrowing room is clean usually occurs after the invested cost of disinfectant and immediately prior to sows entering the room. At the same time, studies have demonstrated that visual inspection of farrowing rooms may be insufficient to ensure cleanliness and reduce disease transmission risk. The goals of this project were to determine the areas of the farrowing crate with the greatest surface contamination, the correlation between microbial counts and relative light units, and the number of locations that are needed to accurately determine surface cleanliness. The results from this study indicated that the areas of highest concern were the entryway floor and the sow feeder as detected both by ATP bioluminescence and coliform plate counts (CPC). Bioluminescence is a monitoring tool that should be used in conjunction with periodic microbial validation to monitor procedures for cleaning and disinfection.
Project #: 22-002 | Principal Investigators: Giovani Trevisan & Daniel Linhares | Institution: Iowa State University | Posted: 7/9/2024 | Keywords: data analysis, surveillance, preparedness, animal health threats, diagnostic
This study evaluated the use of endemic enteric coronaviruses polymerase chain reaction (PCR)-negative testing results as an alternative approach to detect the emergence of animal health threats with similar clinical diseases presentation. This retrospective study, conducted in the United States, used PCR-negative testing results from porcine samples tested
at six veterinary diagnostic laboratories. As a proof of concept, the database was first searched for transmissible gastroenteritis virus (TGEV) negative submissions between January 1st, 2010, through April 29th, 2013, when the first porcine epidemic diarrhea virus (PEDV) case was diagnosed. Secondly, TGEV- and PEDV-negative submissions were used to detect the porcine delta coronavirus (PDCoV) emergence in 2014. Lastly, encountered best detection algorithms were implemented to prospectively monitor the 2023 enteric coronavirus-negative submissions. Time series (weekly TGEV-negative counts) and Seasonal Autoregressive-Integrated Moving-Average (SARIMA) were used to control for outliers, trends, and seasonality. The SARIMA’s fitted and residuals were then subjected to anomaly detection algorithms (EARS, EWMA, CUSUM, Farrington) to identify alarms, defined as weeks of higher TGEV-negativity than what was predicted by models preceding the PEDV emergence. The best-performing detection algorithms had the lowest false alarms (number of alarms detected during the baseline) and highest time to detect (number of weeks between the first alarm and PEDV emergence). The best performing detection algorithms were CUSUM, EWMA, and Farrington flexible using SARIMA fitted values, having a lower false alarm rate and identified alarms 4 to 17 weeks before PEDV and PDCoV emergences. No alarms were identified in the 2023 enteric negative testing results. The negative-based monitoring system functioned in the case of PEDV propagating epidemic and in the presence of a concurrent propagating epidemic with the PDCoV emergence. It demonstrated its applicability as an additional tool for diagnostic data monitoring of emergent pathogens having similar clinical disease as the monitored endemic pathogens.
Project #: 23-031 | Principal Investigators: Daniel C. L. Linhares; Gustavo de Sousa e Silva | Institution: Iowa State University College of Veterinary Medicine | Posted: 7/1/2024 | Keywords: Manure; Manure pumping; PRRS; PEDV; Wean-to-finish; Bioexclusion; Biocontainment
Due to its nutritional and fertilizing value to the soil, the pig manure is spread in fields surrounding pig sites for the following grain crop season. However, manure agitation and spreading poses risks to animal health due to the gases and pathogens that may recirculate in the site and surrounding sites. Therefore, the goal of this project was to estimate the impact of manure pumping practices on PRRSV and PEDV health outcomes in wean-to-finish (W2F) pig populations. A retrospective and prospective studies were conducted separately.
The retrospective epidemiological study estimated the odds of PRRSV or PEDV outbreak occurring within four weeks after manure pumping out from the site event (exposure 1) or being located in a field receiving manure at 1-, 3-, and 5-miles from a site (exposure 2). The study population was W2F lots from one production system that pumped manure between July 2020 and December 2022. PRRSV or PEDV outbreaks (cases) were defined based on veterinarian assessment, pathogen detection in tissues, and increased mortality rate after the pumping event or receiving manure from other site. For the analyses, controls were selected to match spatially (within 6.2 miles of cases) and temporally (placement dates within a 4-week interval from outbreak dates) cases. The analyses revealed that the odds of PRRSV outbreaks events within a 4-week following pumping out of the site and spreading manure activities were higher. Additionally, nurseries had higher odds of reporting a PRRSV outbreak following pumping out activities compared to grow-finish. No associations between PEDV outbreak and manure practices were detected in this retrospective study.
The prospective study assessed the frequency of PRRSV RNA and PEDV RNA detection in pit samples from midwestern W2F barns and the likelihood of increasing PCR-positivity of pig oral fluids after manure pumping. The population of interest was wean-to-finish lots from a swine producer that pumped manure between April 2023 and December 2023. PRRSV and PEDV were tested by PCR on oral fluids and pit manure. All growing pig barns were selected based on the absence of PRRSV or PEDV before the pumping process.
The analyses revealed an increase in the odds of testing PRRSV-positive in oral fluids after pumping out manure. The PEDV positivity in manure was significantly higher than that of PRRSV, however there was no increase in oral fluids PEDV-positivity after pumping manure. Anyways, there was evidence of both PRRSV and PEDV virus in manure samples, confirming the ecological importance of manure in viral spread within and between sites.
Both studies (retrospective and prospective) showed that manure pumping practices were associated with PRRSV outbreak and spread. The odds of a PRRSV outbreak within a 4-week window were greater when the site was pumped and was in close proximity to a field receiving manure. The odds of a previously PRRSV-negative barn becoming PRRSV-positive increased significantly after manure pumping. This information enables veterinarians and producers to justify strategies for biosecurity and biocontainment associated with pumping manure out of sites or when a site is near a field receiving manure.
Project #: 23-037 | Principal Investigators: Daniel Linhares & Edison Magalhães | Institution: Iowa State University | Posted: 6/11/2024 | Keywords: market pigs; trailer sanitation; automated; data integration; compliance verification
The pilot project aimed to address the challenge of documenting truck washes between visits to slaughterhouses and return to swine barns, a critical aspect of market haul sanitation in the swine industry. The primary goal was to assess three different methods for automatically recording truck wash events and market pig deliveries at packing plants, thus, enabling producers to verify trailer cleanliness compliance. Furthermore, automated reports were produced to inform the decision-makers on the status of the trailers, and identify the non-compliance events (i.e., trailers not washed between loads to the packing plant).
The project was conducted in collaboration with one swine producer in the US Midwestern region, focusing on evaluating the feasibility and effectiveness of different technologies in recording truck-related events. Three approaches were tested: GPS tracking of trucks and trailers; a software application (APP) for automatically creating electronic tickets of washing events; and manual data collection at truck wash and packing plant sites. The research team collected and analyzed data over a specified period to evaluate the accuracy and reliability of each method.
The findings revealed that while all three methods had their strengths and limitations, GPS-based tracking showed higher accuracy in documenting truck wash events and deliveries at packing plants compared to the other methods. However, GPS-based methods were susceptible to errors such as false or duplicate events and the geofence limits are not adjusted, highlighting the importance of optimizing technology parameters to minimize discrepancies. On the other hand, despite the CleanTrailer APP having a slightly inferior performance for recording truck wash events, it provided an electronic ticket with pictures of before and after the wash, providing additional information beyond the electronic wash ticket. Despite some missed washes, the agreement between GPS data and the CleanTrailer APP was generally high, indicating the potential of automated systems in ensuring compliance with sanitation standards.
The results of this pilot study have significant implications for the swine industry, as providing producers with automated reports to monitor truck wash compliance. The scalability of the methods tested suggests broader applicability across production systems, offering a standardized approach to monitoring market haul sanitation practices. Ultimately, these findings empower producers to make informed decisions regarding truck sanitation, thereby safeguarding animal health and improving overall industry practices.
Project #: 23-071 | Principal Investigator: Dr. Michael Chetta | Institution: Talent Metrics Consulting | Posted: 5/16/2024 | Keywords: safety, biosecurity, biocontainment, wean-to-market, caretaker, mitigation, prevention, preparedness, compliance
The Swine Health Information Center has identified that caretaker motivation related to compliance with biosecurity behaviors is a priority needing to be better understood. An exploratory study was conducted to establish a baseline for worker motivation and identify possible issues within the industry that could be impacting compliance with biosecurity. This research and measurement related to motivation is the first of its kind in the industry and sets the groundwork for better understanding the primary factors influencing worker motivation and compliance.
Initial findings suggest the swine industry’s problem with biosecurity compliance is not a motivationally driven issue, and not wholly influenced in the way initially conceptualized and measured. There is strong support that biosecurity compliance is influenced by job resources (specifically supervisor support), availability of performance feedback and rewards. Additionally, the analyses suggest workers are heavily impacted in doing their work and adhering to biosecurity protocols by physical workload and demanding contact with animals.
There is reason to believe that motivation can be assessed differently and that the impact of training and measuring the implementation/effectiveness of biosecurity procedures could yield valuable insights. Continuing this research across the industry will help one of the largest industries in the US to better understand the interactions and motivations behind worker attitudes and perceptions towards biosecurity adherence and to enhance positive outcomes for employees, farms, and consumers.
Project #: 23-046 | Principal Investigator: Francisco Cabezon | Institution: Pipestone Research | Posted: 5/16/2024 | Keywords: Power-washing, robotics, cleanliness, water usage, labor
A 2,400 head wean-to-finish barn with two rooms of 1,200 head capacity (196 feet x 50 feet) with 44 pens each was used in the study. A group of nursery pigs were placed in the barn and raised until harvest. The barn was then cleaned, with one room washed using traditional manual power washer methods from a contract service, and the other room cleaned using a railed robotic power washer prototype, followed up with a manual power wash to remove any additional manure (touch-up). The trial consisted of two washing events (August 2023 and February 2024) to compare the efficacy and efficiency of an automated power washer to a manned power-washing crew, based on cleaning time, manpower time, water usage, and cleanliness rate.
In the room washed with the rail robotic power washer prototype, four rails were installed (2 on each side of the room divided by the central hallway) to cover the pen floor and side walls at a maximum height of 10 inches from the slat level. The rail robotic power washer prototype consisted of a trailer head carrying a rotary nozzle connected to a gas power washer. The trailer head was battery powered, and the speed of the trailer on the rail and the speed of rotation of the nozzle could be adjusted. Two different rotary nozzles were tested. The robot power washer with a single rotary nozzle was set to move through the rails at an average speed of 11.0 inches/min, with a nozzle rotation time cycle of 22 seconds (August 2023 data). In the case of the double rotary nozzle, the robotic power washer was set to move at an average speed of 14.8 inches/min, with a nozzle rotation time cycle of 30 seconds (February 2024 data). In both cases, the speed of the trailer head and rotation of the nozzle were adjusted to achieve 2 hits per slat.
Multiple methods were used to evaluate cleanliness (pre-wash, post-wash, and post touch-up): visual assessment, adenosine triphosphate (ATP) measurements to assess organic material, bacterial culture with dip slides, and a reverse-transcriptase real-time PCR (RT-qPCR) for rotavirus detection. There were 12 pens assessed in each room, which were equally spaced throughout the room. Five sites in each pen were assessed: the fencing, floor, wall, waterer, and feeder.
In August 2023 (single rotary nozzle test), total water usage in the robotic power washing room was 8,396 gallons in comparison to 6,211 gallons in the manual power washing room. Total washing time in the robotic power washer room was 22.1 h (13.0 h of robotic washing and 9.1 h of manual touch up washing) in comparison to 10.5 h of manual power washing in the control room. The manual washing labor time in the robotically washed room was reduced 13% (1.4 h), but total washing time was longer by 11.6 h.
In February 2024 (double rotary nozzle data), total water usage in the robotic power washing room was 10,897 gallons in comparison to 7,526 gallons in the manual power washing room. Total washing time in the robotic power washer room was 19.3 h (10.1 h of robotic washing and 9.2 h of manual touch up washing) in comparison to 13.3 h of manual power washing in the control room. In this case, manual washing labor time in the robotically washed room was reduced by 31% (4.1h) with the robot, but overall washing time was longer by 6 h.
Cleaning scores differences before and after washing were significant for each power washer method, at all sites in a pen, and in each testing. The cleanliness trend was from very dirty to clean or very clean. For the robotic power washed room, the post-wash touch-up by the manual power washing team was necessary for the median value to reach the “Very Clean” score.
More bacterial count, rotavirus presence, and ATP levels were found after the washing process for both wash methods. Power washing does not clean the barn, it is solely a means to remove debris and must be followed by a disinfection process. Power washing should be completed to the necessary level to ensure that disinfection can be performed well.
Cleaning expectations of this barn were extremely high, this could explain to some degree the long touch-up process. Robotic power washer cannot easily access the feeders. The washing crew spent considerable time washing the feeders. The number of feeders in the barn will be a limiting factor to the efficiency of the robotic power washer. The barn used for this research has a low pigs/feeder ratio (27 pigs/feeder, doubled 1-hole wet dry feeder). Another limiting factor for the automated power washer is the number of rails and their positioning. In the current study 4 rails were installed in the room. This allowed walls to be washed at a maximum height of 10 inches from the slat level, however, the wash did not cover the central hallway. Additional rails could increase the covered area by the rail power washer, but it would represent additional costs and time of operation.
Although power washing needs at facilities are time and resource intensive, this robotic power-washer prototype does not provide adequate savings in manpower or water usage, so further refinements are needed.
Project #: 23-079 | Principal Investigator: Dr. Mariana Kikuti | Institution: MSHMP/UMN | Posted: 4/17/2024 | Keywords: Porcine deltacoronavirus; epidemiology; swine health; disease occurrence; incidence
PDCoV, an enveloped RNA virus, causes atrophic enteritis in neonatal piglets, leading to diarrhea, malabsorption, dehydration, and death. The study aims to fill the gap in the current epidemiological information about PDCoV in the U.S. pig population after its emergence in 2014. Data from the Morrison Swine Health Monitoring Project (MSHMP) between January 2015 and December 2023 were analyzed, representing approximately 60% of the U.S. breeding herd. Participating herds report weekly PDCoV health status. In total, 244 PDCoV outbreaks occurred in 186 sites from 22 production systems across 16 states. Case counts peaked during winter, and incidence ranged from 0.44% in 2017 to 4.28% in 2023. For sites that experienced more than one PDCoV outbreak during the study period, the interval between outbreaks was a median of 2.11 years. The South and Midwest regions reported the majority of cases. In 2017, a shift in the spatial distribution of cases from the Midwest to the South was observed. The findings underscore the importance of continued monitoring and strengthened control measures to mitigate the impact of PDCoV in U.S. breeding herds.
Project #: 22-059 | Principal Investigator: Dr. Gustavo Machado | Institution: North Carolina State University | Posted: 3/28/2024 | Keywords:Truck Transport, Disease modeling, Contact trace, Indirect contact, Truck cleaning, Disinfection
Project #: 22-076 | Principal Investigator: Dr. Derald J. Holtkamp | Institution: Iowa State University| Posted: 3/27/2024 | Keywords: Biosecurity, hazard analysis, outbreak investigation, investigation form, swine
Sustained progress on improving the intended outcomes of biosecurity has been elusive. Improvement in outcomes for endemic diseases, such as the reduced frequency of outbreaks in sow farms, has not been observed. The rapid spread of PEDV after it was introduced into the US in 2013 and the spread of African swine fever virus (ASFV) worldwide (WOAH, 2023) are reminders of the ongoing threat of emerging and transboundary diseases. A more comprehensive, strategic and deliberate approach to identifying, assessing and prioritizing biosecurity hazards to efficiently allocate biosecurity resources is urgently needed to make sustainable improvements in the intended outcomes of biosecurity. Outbreak investigations are excellent opportunities to improve biosecurity if the aim of the outbreak investigation is to collect enough information to confidently identify, analyze and prioritize the biosecurity hazards that need to be addressed first.
In 2021, the Swine Health Information Center (SHIC) funded the development of a Standardized Outbreak Investigation Program. A working group of fourteen swine veterinarians was formed to develop terminology, the approach and an investigation form and reports for conducting outbreak investigations and reporting the findings. The Standardized Outbreak Investigation Program borrowed heavily from methods developed for the hazard analysis and critical control point (HACCP) methodology and may be used to conduct investigations on sow farms as an integrated biosecurity hazard analysis and epidemiological investigation.
A critical concept for identifying biosecurity hazards is the three failures. This concept is foundational for the standardized outbreak investigation conducted as a biosecurity hazard analysis. The three failures are 1) failure to prevent the pathogen-carrying agent from being contaminated or infected with an infectious pathogen, 2) failure to mitigate the contamination or infection of the pathogen-carrying agent, and 3) failure to prevent pig(s) in the herd from being infected with the pathogen from the pathogen-carrying agent after the pathogen-carrying agent arrives at the site. All three failures are necessary for transmission to occur. Using the three failures concept, a biosecurity hazard can be defined as any aspect of the production process that increases the probability of one or more failures. A biosecurity hazard analysis is conducted by investigating the production procedures and the pathogen-carrying agents associated with entry events to identify the biosecurity hazards.
The form developed for the Standardized Outbreak Investigation Program was originally formatted as a Word® (Microsoft, Redmond Washington) document. The limitations of the form formatted as a Word document were that 1) it was relatively difficult to use, 2) there was not a convenient way to store and access company-specific historical information collected during investigations, and 3) it was not feasible to capture the data from all users to build a secure industry-wide database that could be used to learn from the collective experience of veterinarians and producers. The aim of this project was to develop a web-based application and database for the Standardized Outbreak Investigation Program to overcome those limitations.
The Standardized Outbreak Investigation Program web application may be accessed by producers or veterinarians at https://fieldepi.org/soip/. Requests to set up a new company may be made by email to [email protected]. Companies and sites can be created in the application and investigations for several swine pathogens may be conducted for individual sites. The application includes an easy to navigate form to conduct the investigations. The form is organized into two main sections. The first section is the pre-investigation survey. It is intended to capture information before the investigation is conducted on-farm. The second section is the investigation form which is to be used to capture information during the investigation conducted on-farm and from any follow-up conducted.
Upon completion of an investigation, a detailed report and an executive summary may be viewed and saved as PDF files. The detailed investigation report includes all the information collected from the pre-investigation survey and the investigation form. The executive summary is a shorter report that summarizes the investigation’s major findings, including a table of hazard ratings for each entry event and the frequency of each entry event during the investigation period. A summary of the entry events that were rated high, including the justification for rating the event as high, is included in the executive summary. The executive summary also includes a summary of the weather during the investigation period.
Outbreak investigations are magnificent opportunities to identify and prioritize biosecurity hazards. Outbreaks are a crisis and an opportunity to learn, but learning is not guaranteed. If outbreak investigations are conducted systematically, deliberately and comprehensively to identify, assess and prioritize biosecurity hazards, the information learned will provide a return on the investment in the investigation and sustained progress can be made on the intended outcomes of biosecurity.
Project #: 22-072 | Principal Investigator: Dr. Natalia Cernicchiaro | Institution: Kansas State University| Posted: 3/22/2024 | Keywords: Japanese encephalitis, JE, JEV, risk assessment, swine
Japanese encephalitis virus (JEV), which is associated with encephalitis in humans, and reproductive and neurological illness in pigs, has expanded to areas free from disease, most recently in Australia with the invasion and expansion of JEV in eastern and southeastern states. The current study’s overarching goal is to increase awareness and build capacity to cope with the risk of a potential JEV incursion. The introduction of this virus into the United States (US) could not only impact public health, but also animal health and the food supply. To efficiently and cost-effectively manage risk, a better understanding of how and where this disease will be introduced, transmitted and spread is required. The objective of this study was to update and reassess the 2018 qualitative risk assessment implemented to estimate the risk of emergence of JEV into continental US and add information regarding transmission post-emergence, establishment and spread, by incorporating the latest scientific information, newer assumptions, data sources, recent outbreak data from Australia, and regional elements contributing to the risk, which may have been previously overlooked.
We used an established semi-quantitative risk assessment framework to evaluate the risk of introduction of JEV into seven US regions from the current region of distribution (i.e., southeast Asia and the western Pacific rim), and its subsequent spread and consequences. The regions at risk were created by grouping the 48 states of the contiguous US into five regions based on similar climate, density of domestic swine production, and presence of feral swine; due to their geographic isolation, Alaska and Hawaii were included as their own individual regions. Information utilized to update model parameters included data from an updated systematic review of the literature on vector and host competence for JEV. Our updated review assessing literature published from 2016-2022 reported a slightly higher number of articles that focused on host competence and susceptibility to JEV compared to vector competence. This shift in focus could be attributed to, in part, the realization of certain host species’ (e.g., pigs, migratory and domestic birds) high susceptibility to JEV. One striking feature explored in an experimental study, which has yet to be replicated, is the ability of JEV to be transmitted between pigs without the aid of mosquito vectors. Consequently, there is a growing recognition of the importance of controlling not only the vector, but also the virus at the animal level, particularly through vaccination and other control strategies.
Additional sources of information for our risk assessment models included global databases, scientific literature, and expert opinion obtained from our advisory group and project collaborators. Our advisory group consisted of swine producers, veterinarians, entomologists, researchers and stakeholders from the US and Australia with knowledge on commercial swine production, Japanese encephalitis virus, and the latest Australian outbreak.
Summary output parameters calculated by this risk assessment include the rate of introduction, estimated epidemic size, and the overall risk estimate. The highest risk scores for the rate of introduction were deemed moderate and associated with the entry of adult mosquitoes in cargo ships into the US south (RAR 3; AL, AK, DE, Fl, GA, KY, LA, MD, MS, NC, OK, SC, TN, TX, VA) and west (RAR 5; CA, ID, OR, WA) regions, and via eggs/larvae in imported tires into the northeast (RAR 1; ME, MA, NH, NY, VT) and west (RAR 5) regions. The largest estimated epidemic size was greater than one million host animals in the US south (RAR 3), whereas the lowest estimated epidemic size was 10 host animals in the northeast (RAR 1). In the midwest region (RAR 2; CT, IL, IA, KS, MI, MN, MO, NE, NJ, ND, OH, PA, RI, SD, WV, WI) the estimated epidemic size ranged from 10 to between 1,000 and 10,000 host animals, and in the west (RAR 5) region from 100 to between 100,000 and one million. The overall risk, which reflects the rate of introduction and economic impact of a JEV incursion into the region at risk, was very high for the south (RAR 3), moderate for the west (RAR 5), and very low for the northeast (RAR 1) and the midwest (RAR 2) regions.
In summary, based on the results from this risk assessment, the US south region (AL, AK, DE, Fl, GA, KY, LA, MD, MS, NC, OK, SC, TN, TX, VA) should be prioritized for preparedness to mitigate the impact of disease, whereas the west (CA, ID, OR, WA) and northeast (ME, MA, NH, NY, VT) should also be prioritized for surveillance activities towards early disease detection.
As with any risk model, the results of this study are entirely dependent on the data used and the assumptions made. In this assessment, we supplemented the limited data on JEV transmission (e.g., the role of vector-free transmission among pigs and of non-ardeid birds and wildlife in the JEV transmission cycle in US), spread, and persistence, with information gathered from the latest Australian JEV outbreak of 2021-23, existing and new available scientific evidence, and expert opinion. This qualitative risk assessment provides a framework necessary for decision-makers to assess the risk of JEV introduction and potential impacts in the US, which is crucial to prepare for the threat of an emerging disease and protect the swine industry from suffering huge economic losses.
Project #: 22-094 | Principal Investigator: Dr. Cesar A Corzo | Institution: University of Minnesota | Posted: 3/20/2024 | Keywords: Morrison Swine Health Monitoring Project, MSHMP, disease monitoring, PRRS, PEDV, PDCoV, Senecavirus
Objective 1: Monitor trends in pathogens incidence and prevalence – PRRSv, PEDv, PDCoV, Senecavirus and central nervous system associated viruses continued to be monitored and each maintained the historical pattern. We also investigated the relationship between the PRRS RT-PCR positivity rate between breeding and growing pigs finding a correlation; however, because no spatial information and prrs virus information was accounted for we need to interpret this information with caution, thus we are unable to conclude that the increase in PRRS positive rate in breeding herds is due to an increase in positivity rate in growing pigs. In addition, we also assessed the frequency of PRRS outbreaks in breeding herds managing mortality either through composting, incineration, or rendering. Numerical differences were detected but not enough evidence was found to conclude that one method is better than the other one and more analyses are needed. Regarding PEDV, we characterized the time positive herds required to stabilize the herd and concluded that those infected during the epidemic phase of the disease in the US took longer (e.g., 9 weeks) to reach stability than those infected during the endemic phase.
Objective 2: To conduct prospective monitoring of PRRSv sequence evolution and impact – We characterized the epidemiological situation of an emerging virus (L1C-124) that was thought to be a virulent and fast spreading one. Even though the virus did disseminate, it did not reach a speed and magnitude of other recent viruses. The clinical impact was also characterized but when compared to the recent L1C-144 we can conclude that it was not significantly different.
Objective 3: To expand participation of producers to allow for all to be involved – During 2022-2023 we have added 4 production systems. Our database now has 1,274 sow farms housing approximately 3.8M sows, 3,318 growing pig farms with 12.2M pig spaces and 48 boar studs totaling 12,799 boars. Our first publicly available MSHMP website has been completed, officially launched in Q3 of 2023, and has been continuously updated. This website was designed to optimize MSHMP output dissemination within both the participant and industry communities. The website represents a significant milestone in our mission to enhance collaboration and communication within the swine industry. The website has been designed with ten comprehensive sections, including Home, About, History, People, Reports, Outreach, Ongoing Projects, News, Resources, and Contact Us. The website has been accessed by individuals from 41 countries with the highest number of users from the US, China and Spain.
Project #: 23-045 | Principal Investigator: Dr. Rahul K. Nelli | Institution: Iowa State University | Posted: 3/20/2024 | Keywords: Japanese encephalitis virus, JEV, RT rt-PCR, JEV detection
Japanese Encephalitis Virus (JEV) is a severe disease affecting pigs and humans. It is spread mainly by mosquitoes and is endemic in many parts of Asia and the western Pacific regions. However, its recent outbreak in Australia posed a renewed threat to the US pig industry and public health. Therefore, it is crucial to have a reliable way to detect JEV in pigs before it causes an outbreak. The current project aims to detect JEV rapidly using viral RNA and Reverse Transcription real-time polymerase chain reaction (RT-rtPCR) assays. This project established RT-rtPCR assays for JEV and its five genotypes (G-1, -2, -3, -4, -5) and compared the newly established test with a previously published assay. The findings from this project suggest that the newly developed RT-rtPCR assay is more specific and can detect all five genotypes accurately and efficiently. It also had a low detection limit, meaning it could detect very small amounts of the virus. Validation of genotype-specific RT-rt PCR assay and evaluation of the assay performance on known status clinical samples in collaboration with Australia is ongoing. Our project developed a novel and reliable test for JEV that can help the pig industry and public health authorities monitor and prevent JEV outbreaks.
Project #: 22-065 | Principal Investigators: Marcelo Almeida, Alyona Michael | Institution: Iowa State University | Posted: 2/5/2024 | Keywords: App, surveillance, oral fluids, nasal swabs, tonsil scrapings, survivability
After an outbreak of Actinobacillus pleuropneumoniae (App) serotype 15 in central IA in 2021/22 three initiatives were put in place to investigate the atypical behavior of this event. Serological surveillance was performed in 19 of 30 sow farms supplying pigs to sites experiencing outbreaks. Most sow farms (16 of 19) were App antibody negative, suggesting that at least part of the finishing sites may have had a lateral introduction of the bacteria. Evaluation of individual (nasal swabs and tonsil scrapings), population (oral fluids) and environmental samples were collected longitudinally over 6 weeks starting 3 weeks post the reported App 15 outbreak in a finisher to evaluate post-outbreak persistence dynamics. Tonsil scraping had the highest detection rate of App by PCR, while nasal swabs only detected positive pigs in the first 3 weeks of sampling. Oral fluids could detect positive pens during the 6 weeks, but missed pens with positive pigs. Environmental samples were sporadically positive by PCR. Finally, a comparative evaluation of three different App strains (serotype 15 outbreak and historical strain and serotype 1) were compared in liquid media and solid surfaces at different temperatures for up to 7 days. Warmers temperatures (25°C and 37°C) decreased the survivability of all three strains and colder temperatures (-20°C and 4°C) were favorable to survivability. Differences were observed in liquid media (higher survivability in horse serum-NAD compared to water and fecal slurry) and solid surfaces (concrete > rubber > stainless steel). This information adds to App ecology and epidemiology knowledge and may help inform about diagnostic, monitoring and surveillance, and biosecurity decisions.
Project #: 21-133 | Principal Investigator: Chad Paulk | Institution: Kansas State University | Posted: 12/19/2023 | Keywords: Chlorine Dioxide, Feed Mill Decontamination, Flushing, Formaldehyde, Viral contamination, Paulk
This project set forth to understand the best flushing, thermal processing, and decontamination techniques for a feed mill contaminated with porcine epidemic diarrhea virus (PEDV), porcine reproductive and respiratory syndrome virus (PRRSV), and Seneca Valley virus 1 (SVV1); furthermore, infectivity of samples collected after utilizing these techniques was evaluated using a swine bioassay. Formaldehyde flushes (either liquid or dry) were most effective at reducing viral concentrations in both the feed and the environment; however, no method eliminated viral RNA from the environment. Although PEDV and SVV1 RNA were detected via PCR, upon inoculation to pigs, treatments failed to cause infection which provides evidence of successful mitigation. However, PRRSV viral particles were able to cause replication in pigs when introduced from both feed and dust samples even when the inoculum samples were negative for PRRSV via PCR. It is unknown why PRRSV RNA was not consistently detectable via PCR, but the bioassay suggests more research is needed, especially regarding PRRSV mitigation. Thermal processing reduced the detectable RNA in both feed and the environment for all viruses. Replication of SVV1 and PEDV was not observed in the swine bioassay, but PRRSV replication was observed in the post-thermal processing feed. Increased temperatures may be required to inactivate PRRSV if contamination were to occur pre-thermal processing. The complete facility decontamination (removal of organic matter with heated pressure washing, disinfection with 1% Virkon, disinfection with 5% household bleach, environmental heat held at 140°F for 48 hours) was the only decontamination treatment where PEDV, PRRSV, and SVV1 RNA was non-detectable after completion of all steps. The chlorine dioxide and heat treatments reduced the detectable quantity of RNA, but viral particles were still detectable across mill surfaces. Although PEDV and SVV1 RNA was detected via PCR, only PRRSV replication was observed in the swine bioassay from the chlorine dioxide treatments and one heat treatment. Overall, chemical flushing, thermal processing, and facility decontamination reduce the viral RNA presence, but more research is needed to understand how these techniques affect the infectivity of these viruses.
Project #: 23-018 | Principal Investigator: Dustin Boler, Bailey Harsh | Institution: Carthage Innovative Swine Solutions, University of Illinois | Posted: 11/14/2023 | Keywords: Truck wash biosecurity, ATP bioluminescence, disease monitoring, surveillance
This research evaluated the performance of two adenosine triphosphate (ATP) instruments to estimate cleanliness in livestock trailers. The results of the ATP instruments were compared to aerobic plate counts (bacterial contamination) to determine if ATP bioluminescence could be used as an indicator of trailer cleanliness without the need for visual inspection. ATP is a source of cellular energy that is present among all living organisms. This includes viruses and bacteria that remain after a commercial trailer cleaning. Normally, trailer cleanliness is determined visual evaluation by a person that inspects the trailer to determine if it is free of organic material and suitable to return to a farm. However, studies have demonstrated that visual inspection of cleaned transport trailers may be insufficient to ensure cleanliness and reduce disease transmission risk because viruses and bacteria are microscopic in nature and cannot be seen by the human eye. Further, visual inspection to determine if a trailer is clean usually occurs after the invested cost of propane to dry the truck has occurred. ATP bioluminescence uses a chemical reaction where a swab is used to detect the presence of ATP. The more ATP that is present, the greater the chemical reaction. This technology uses the same chemical process a firefly uses to illuminate. When ATP is exposed to the enzyme a light is produced. The more ATP that is present the brighter the light. The intensity (brightness of the light) is measured in relative light units (RLU). A greater RLU indicates more ATP and reduction in overall cleanliness. So, this technology can be equated to the brightness of a firefly. The brighter a firefly glows; the more ATP is present. In this case, the brighter the swab glows, the more ATP is present, the more potential microbial contamination is present. The goals of this project were to determine the areas of the trailer with the greatest surface contamination, the correlation between microbial counts and RLUs, and the number of locations that need to accurately determine surface cleanliness.
This project evaluated livestock trailers at two commercial trailer washes. One of those locations included trailers that were known to transport livestock from a farm known to be PRRS or PEDv positive. In total, 100 livestock trailers were tested usings biolumiomters to determine the amount of ATP that remained after a commercial trailer wash. Protocols are in place to prevent people from entering a livestock trailer after it has been cleaned to prevent contamination. For this technology to be adopted into practice, locations inside the trailer that are accessible from outside the trailer must be evaluated. This trial evaluated the back door flush gate (BDFG), rear drivers side access door (RSAD), the belly flush gate (BFG), belly side access door (BSAD), nose side access door (NAD).

Figure 1 Back door flush gate. An area of the trailer that is indicative of overall trailer cleanliness.
The results from this study indicated that the areas of highest concern sampled in this study were the nose access door and the back door flush gate as detected both by ATP bioluminescence and APC. A key finding of this research was that nose access door was the area least likely to be adequately cleaned, but only a few trailers actually had nose access doors. Nearly all of the trailers evaluated had a back door flush gate and therefore makes it a logical place to swab a livestock trailer to determine the overall cleanliness.
The ability of ATP to be used as an indicator of trailer cleanliness was dependent on the instrument used, with the 3M machine being more closely correlated with bacterial contamination. Swabs were also collected to determine the presence of PEDv, but all swabs were negative. These data suggest that ATP bioluminometers can be used in livestock trailers to quickly determine the general cleanliness of the trailer without the need for a visual inspection. Bacterial swabs to determine APC levels should also be used to determine the effectiveness of the cleaning protocol.
Project #: 22-071 | Principal Investigator: Dr. Monteserrat Torremorell | Institution: University of Minnesota | Posted: 9/12/2023 | Keywords: whole-genome, long-read sequence, viruses, target enrichment
Viral co-infections on swine farms are common and frequent, contributing to aggravated disease outcomes and impairing economic profit and animal well-being. In addition, due to high mutation rates, the co-circulation of viruses within swine herds facilitates the emergence and re-emergence of new variants, impairing disease investigation, control, and prevention. Despite their significant cost and health relevance, viral co-infections are understudied in part due to limitations in sequencing techniques able to generate full-length sequences of multiple viruses from field samples. Furthermore, the detection and genomic characterization of novel variants are still a challenge, and many questions remain about the dynamics and interactions of co-circulating viruses in swine herds. To advance our understanding of viral co-infections, we have developed a workflow called “TELSVirus”, or “Target-Enriched Long-read Sequencing of Virus” that enables the real-time detection and genomic characterization of multiple viral pathogens from a single sample in a relatively short turnaround time (~24 h). The main objective of this study was to apply the “TELSVirus” workflow to porcine oral fluid samples to detect and characterize genomes of target viral pathogens. Overall, this method allowed us to detect a high prevalence of co-circulating yet understudied viruses, including porcine bocavirus, porcine sapelovirus 1, and porcine astrovirus 2 and 4; while also allowing for detection of viruses with known production impacts, such as porcine reproductive and respiratory syndrome virus (PRRSV), influenza A virus (IAV) and rotavirus. These viruses were often detected in the same oral fluid sample, and the high sequencing coverage afforded by TELSVirus allowed for robust variant analysis. We also demonstrated that TELSVirus’ limit of detection for PRRSV and IAV is comparable to that of qPCR, while providing increased genomic information. Based on these results, TELSVirus has the potential to support real-time surveillance of endemic and emergent viruses, while also improving our understanding of co-circulating viruses; their genetic diversity; and ultimately how they impact swine health and production.
Project #: 21-139 | Principal Investigator: Pablo Pineyro | Co-investigator: Marcelo Almeida | Institution: Iowa State University | Posted: 8/28/2023
Porcine circovirus type 3 (PCV3) was initially described as affecting swine in 2016, and since its first description, PCV3 has been detected globally and retrospectively in cases of pigs of all ages. It has been associated with a range of clinical presentations, including reproductive failure, porcine dermatitis and nephropathy syndrome, multisystemic inflammation, respiratory disease, enteric disease, and subclinical infection. However, for many of these clinical presentations, the causative role of PCV3 remains questionable, and only a few are real clinical problems. The current diagnosis is based on viral detection by PCR and occasional in situ confirmation by in situ hybridization. However, PCV3 has been detected in clinical and subclinically infected animals. Therefore the lack of correlation between the viral detection and the presence of lesion(s) or specific clinical signs make it imperative to redefine and improve the PCV3 case definition that can be used to trigger field investigation and better control the PCV3 infection.
One of the objectives of this study was to establish a Ct PCR value that correlates with the presence of lesions compatible with PCV3 infection. Our retrospective study showed that there is a positive correlation between Ct values and the presence of lesions. Cases confirmed by histopathology with lesions consistent with multisystemic inflammation showed Ct values below 30. In addition, the presence of PCV3 in these cases was confirmed by in situ hybridization, adding diagnostic value to the combination of histopathology and PCR detection.
In this study, the evaluation of samples that could be used for surveillance or subclinical detection included oral fluids, processing fluids, and serum. Most of these samples were not linked to specific clinical problems. Evaluation of the Ct value distribution demonstrates that a high proportion of the sample presents Ct values above 32. The implication of these findings needs further evaluation. These sample matrices could indicate viral shedding, vertical transmission on sow units, or viremia in grower-finisher. However, the clinical implication of positive results on these sample matrices cannot be extrapolated to assess the presence of PCV3-associated disease.
The evaluation of submissions of reproductive failure associated with PCV3 showed that Ct values on cases confirmed only by PCR or a combination of PCR and histological evaluation do not differ. These results suggest that PCR on fetal tissue could be sufficient to confirm causation. However, a high proportion of cases showed coinfection with PRRSV and PCV2. The Ct values on these coinfection cases showed a positive correlation suggesting that these viruses have a synergist effect with a higher PCV3 viral load when found in association with PRRSV and PCV2.
We conclude that the diagnosis of PCV3-associated disease should be based on a combination of diagnostic tools, including lesions of multisystemic inflammation, significant PCR Ct values lower than 30, and additional confirmation by direct detection methods. The sample used for surveillance and subclinical evaluation could be used as a proxy for viral circulation in sow herd and grower pigs but cannot be correlated with multisystemic inflammation or reproductive failure.
Institution: University of Minnesota | Posted: 7/2019
Vitamins are essential nutrients required by swine to optimize health, productivity and well-being. The U.S. pork industry is dependent on vitamins manufactured in China because there are limited, and in some cases, there are no other country of origin options to meet industry volume demands. Initial studies have provided emerging evidence that the African Swine Fever virus (ASFv) can survive in choline chloride, but not vitamin D3. However, it is unknown if this virus can survive in other vitamins. The risk of ASFv or other Foreign Animal Diseases (FAD) being introduced from China into the U.S. through vitamin imports appears to be low, but the impact of introduction is high.
Project #: 22-070 | Principal Investigator: Dr. Natalia Cernicchiaro | Institution: Kansas State University | Posted: 7/14/2023 | Keywords: swine, Japanese encephalitis, rapid systemic review, transmission
The United States (US) is considered a susceptible region with great potential for the introduction of the Japanese encephalitis virus (JEV) given the availability of competent mosquito vector species (e.g., Culex, Aedes spp.), susceptible maintenance avian hosts (e.g., ardeid, water birds), intensive travel and trade activities to and from Japanese encephalitis (JE)-affected countries, similar climatic and environmental conditions to epidemic countries, no active JEV surveillance in place, and large populations of susceptible swine which can serve as amplification hosts. Of note, is the considerable geographical expansion shown by the JEV in the last decades, including the recent emergence of a new genotype in the eastern and southeastern Australian states. The ongoing risk of the emergence of JEV underlines a need for increased understanding of the biology of this virus, components and dynamics of transmission, and the environmental factors necessary for incursion and establishment.
With funds provided by the Swine Health Information Center (SHIC), a team of researchers at the College of Veterinary Medicine, Kansas State University, led by Dr. Natalia Cernicchiaro, associate professor of veterinary epidemiology, and in collaboration with researchers at the United States Department of Agriculture, National Bio and Agro-Defense facility and the National Feral Swine Damage Management Program, appraised scientific literature to synthesize existing evidence on the role of domestic and feral swine in the transmission of the JEV, and to identify knowledge gaps that can pinpoint areas amenable for future research. From an initial 3,183 article abstracts obtained from electronic databases and hand searches of pertinent literature, a total of 228 articles were deemed relevant and their data extracted for inclusion in this review.
Typically, JEV infection is initiated following the bite of an infected mosquito, although recent evidence indicates that direct contact between pigs may also play a significant role in transmission. Although vector-borne remains the most efficient means of transmission, recent findings indicate that direct pig-to-pig transmission may occur primarily via oronasal fluids, yet additional research is needed to ensure replication of those findings and further evaluate its implications for pathogenesis and spread. Considering vectored transmission, JEV replicates in the skin and reaches the circulatory system through the lymphatic system. In the blood, the virus can be detected as early as 1-day post-infection (dpi) and can persist for 4 to 5 days, regardless of the portal of entry. Virus titers can reach concentrations of 105 50% tissue culture infectious dose/mL (or 106 plaque forming units/mL) between 1- and 5-dpi. Once pigs are exposed to the virus, on average it takes 3 to 6 days, and up to 21 days, to observe clinical signs. Amplification and persistence mechanisms, as well as the effect of genotype in infectiousness, transmission and pathogenesis in swine need to be better elucidated.
Disease is generally mild with a neurologic or reproductive presentation depending on the age and sex of the pigs. Although maternal antibodies can confer protection for longer than four months under field conditions, naïve piglets can manifest neurologic signs including ataxia and tremors which can progress to wasting disease or mortality. Reproductive disorders including stillbirths, abortions, mummified fetuses, and delayed farrowing can occur when sows are infected before 60 to 70 days of gestation. Orchitis, reduced sperm motility and temporary infertility have been reported in boars.
Serological testing followed by virus isolation for confirmation is recommended for diagnosis. During outbreak investigations, serial collection of serum samples in pigs is used to display an increasing titer against JEV. Different tissues are amenable for virus isolation including spleen, liver, brain, sera, cerebrospinal fluid, tonsils, and oronasal fluids. In fetuses, stillbirths or neonates, virus may be isolated or detected from brain, tonsil, spleen, placenta, thoracic or abdominal stillborn fluids. Serologic tests that demonstrate antibodies in fetuses also are useful in diagnosis.
Given no specific therapy is available for treating JEV infection in pigs, control either via mitigation of reproductive losses by vaccination or application of biosecurity practices is critical. Articles describing biosecurity practices for control and risk management at the farm level were very limited. That includes the application and effectiveness of mosquito control and other pest management practices. Vaccination against JEV is the most effective measure for controlling JEV in humans and pigs. Although there are no licensed JEV vaccines for pigs in the US, live attenuated vaccines have shown higher efficacy than inactivated vaccines in natural infection and challenge studies. Vaccinating the breeding stock can control viral amplification and reproductive disease in swine. Before the mosquito season starts, young gilts and boars can be vaccinated twice with a 14 to 21 days interval. In endemic areas, growing pigs are also often vaccinated.
Evidence on risk factors for JEV morbidity pertaining to pigs and their production environment, genetic, sex, and breed differences, as well as management practices at the farm, barn or animal levels, among others, were infrequent to not existent. Although some studies reported the use of pigs as sentinels of infection, the design, implementation and effectiveness of surveillance programs in pigs was rarely described. Even though no peer-reviewed articles were retrieved on preparedness and response efforts related to pigs, national response plans for the animal and human health sectors from governments or other official entities are available from some countries where JEV is present. Of note, are all the resources and educational materials produced in response to the most recent JEV outbreaks by the Australian Government, Department of Health and Aged Care, including the Joint National JEV Outbreak response plan published in June 2023 (JEV outbreak plan). Economic assessments associated with swine, either domestic or feral, and JEV transmission, at the regional, country, state or farm levels were not identified with our search. Similarly, articles discussing impacts or implications of JE in pigs or swine operations were not available.
Pigs have a unique role in the JEV transmission cycle. Not only are pigs implicated in sustaining transmission and spillover to other hosts, but they can also exhibit clinical signs of reproductive and neurological disease, and, although not confirmed in natural infection studies yet, can develop persistent infection of their lymphoid and nervous tissues. Knowledge of the dynamics of the disease process, viral transmission mode, portals of entry and exit, control and treatment options, as well as potential impacts of the disease in swine production, which were appraised by this team and provided in this review, may inform researchers, stakeholders, and policy makers on effort prioritization and development of preparedness and response measures.
Project #: 20-174 | Principal Investigator: Maria Sol Perez Aguirreburualde | Institution: University of Minnesota | Posted: 12/21/22 | Keywords: swine diseases, global surveillance, hazards, preparedness, awareness
Since 2018, the continuous expansion of ASF through Asia, Europe, and the Americas has kept raising concerns in the industry, resonating with the events of 2013, when a devastating epidemic of porcine epidemic diarrhea (PED) virus caused far-reaching losses. Those events demonstrated the importance of developing systems to provide situational awareness to stakeholders in near-real time to coordinate actions between government agencies and the industry with the ultimate objective of preventing or at least mitigating the impact of disease epidemics. The swine industry is vulnerable to the introduction of pathogens, and their variants, from which the US is currently free. We have developed a private- public-academic partnership to support a system for near real-time identification of hazards that will contribute to the mission of assessing risks to the industry. Identified hazards were shared monthly with swine practitioners and the government to help increase the country’s awareness and preparedness. Ultimately, the system has kept contributing to identifying and early detection or creating awareness of key stakeholders to support current prevention and mitigation strategies for the introduction of foreign pathogens into the US.
Project #: 21-109 | Principal Investigator: Montserrat Torremorell | Institution: University of Minnesota | Posted: 12/1/2022 | Keywords: biosecurity, biocontainment, swine, technologies, airborne
Among all infectious agents, airborne pathogens are the most difficult to control and in today’s agricultural settings, airborne animal diseases are the most difficult to contain. This is especially relevant for swine reared in confinement and in controlled ventilated environments where exhaust fans help transport particles from inside to outside of the facility. We conducted a review of technologies directed at removing or/and inactivating pathogens for their application to swine farms and found that only air filtration solely consisting of fibrous filters is an established and implemented technology. There are also industrially and medically implementable technologies named electrostatic precipitators and ultraviolet-C systems (both in-duct and upper room), that they are commonly implemented in other industries but are less prevalent in livestock management. Nonetheless, because they have demonstrated scalable performance in aerosol particle removal and/or bioaerosol inactivation, they merit discussion and consideration towards agricultural biosecurity. Lastly, there are emerging technologies, consisting of more recently developed technologies, typically at the laboratory or bench scale that have not been tested at the scales (building sizes and flow rates) required for agricultural biosecurity and are hence several developmental steps away from direct implementation. Lastly, there are protocols and practices ready to be used at the farm level (e.g. wind breaks, covering of fans, movement of infected animals to other sites, depopulation and reduced ventilation) that conceptually, could be employed in the face of a disease outbreak, but their direct impact on reducing the emission airborne pathogens needs to be evaluated and quantified.
Project #: 21-116 | Principal Investigator: John M. Drake | Institution: University of Georgia | Posted: 9/15/2022
A collaboration between SHIC and the Center for the Ecology of Infectious Diseases (CEID) at the University of Georgia examined spillover risk of bacteria from wild North American mammal species into the U.S. swine herd.
The first phase of the project consisted of a data-driven horizon scan which identified 102 bacteria species hosted by 127 North American wild mammal species that have a measurable chance of associating with domestic pigs. These bacteria species were further analyzed and assigned a propensity score based on their predicted ability of spilling over from their wild mammal hosts into domestic swine. A classification of either novel or known was also assigned based on whether or not these species had a known association with pigs documented in academic literature.
The second phase of the project involved a qualitative approach through the use of a survey of subject matter experts (SMEs). The goal of the survey was to rank high-risk bacteria species to assess their potential impact on the swine industry. Five industry professionals completed the survey and identified 16 bacteria species as being of high impact to the swine industry, four of which are not yet known to infect domestic swine: Anaplasma bovis, Clostridium botulinum, Klebsiella pneumoniae, and Yersinia pestis. Additionally, 7 bacteria were ranked as having a high-risk of antimicrobial resistance, and 5 bacteria were ranked as having a high human outbreak potential.
Future steps involve continued efforts to investigate pathways for potential spillover events, such as shared habitats between wildlife and domestic swine, or pathogen overlap. Additionally, different bacterial strains, serovars, and variants of bacteria species of concern must be closely examined to increase predictive power. Bacteria account for a high degree of morbidity and mortality in swine, causing huge losses to the swine industry and zoonotic risks. Domestic pigs often acquire these pathogens (either directly or indirectly) from wild animals, creating the need for assessments such as these to prioritize bacteria species for targeted prevention strategies. Industry professionals across disciplines hold valuable insights crucial to informing risks related to emerging diseases. Their collective knowledge based on years of expertise fills a gap in the absence of a comprehensive database on the pathogenicity of swine-associated bacteria. A lack of published academic literature also fuels the need to engage SMEs to develop a systematic process for identifying bacterial species of concern to the swine industry, and this research represents a summary of such an endeavor.
The benefit of this collaboration to the industry is that bacteriologists, diagnosticians, and other SMEs will have enhanced information needed to prevent, prepare, and respond to emerging diseases and their potential impact on swine health, welfare, and market.
Project #: 21-113 | Principal Investigator: Gustavo Silva, DVM, MS, PhD | Institution: Iowa State University | Posted: 9/15/2022 | Keywords: PRRSV, PEDV, biosecurity, temperature, time
Decontamination of fomites coming into sow farms using foggers at supply entry rooms is commonly used to mitigate associated risks. Although this practice has been established, recent research has begun to question the effect of this method to inactivate pathogens, especially in complex situations where pathogens may be shielded by organic material or blind spots (Kettelkamp et al., 2019; Leuck et al., 2020).
Objective: The primary objective of this study is to evaluate the efficacy of temperature and time for inactivating porcine reproductive and respiratory syndrome virus (PRRSV) and porcine epidemic diarrhea virus (PEDV) on experimentally contaminated surfaces commonly found at supply entry rooms in swine farms.
Materials and methods: The PRRSV MN184, PRRSV 144 L1C variant or PEDV Viruses were used. The surfaces were diamond plate aluminum and cardboard tested at four temperatures (68 ºF, 86 ºF, 104 ºF and 122 ºF) in combination with six holding times (15 minutes, 60 minutes, 6 hours, 12 hours, 24 hours, and 36 hours). Once the surface temperature reached the desired temperature, the coupons were held for the designated holding time. The negative controls remained at room temperature for 36 hours and the positive controls remained at room temperature for 15 minutes. Three replicates for each treatment were performed and each coupon was inoculated with 2 mL of virus or 2 mL of media for negative controls. Virus titration was performed in each sample separately after the holding time. Regression models and Weibull curves were built to assess the impact of temperature and time on virus inactivation.
Discussion: Under the conditions of this study, PRRSV 144 L1C variant was inactivated from aluminum surfaces by heating coupons to 86oF for 12 hours and for cardboard surfaces by heating the coupons to 86oF for 6 hours. Regarding PRRSV MN184, virus inactivation was possible at 86oF after 24 hours on aluminum and at 104oF after 12 hours on cardboard. PEDV inactivation was achieved at 86oF after 6 hours on aluminum and at 86oF after 12 hours on cardboard. Virus was inactivated after 15 minutes and 1 hour at 122oF in aluminum surfaces for PEDV and PRRSV 144 L1C variant, but not for cardboard.
Overall, regardless of the virus tested on this study, to reduce the risk of virus introduction at swine farms through a supply entry room, it is recommended that materials should achieve 860F, and remain at that temperature for 24 hours. Other options, would be to increase the temperate to 104oF or 122oF but decreasing the holding time for 12 hours.
Project #: 20-078 | Principal Investigator: Hiep Vu | Institution: University of Nebraska-Lincoln | Posted: 7/1/2022 | Keywords: ASFV, ASF, pen-side test, lateral flow, PCR, early detection
African swine fever (ASF) is a devastating viral disease of domestic pigs with mortality rate that can approach 100%. ASF control mainly relies on strict biosecurity that involves movement restriction, quarantine, and compulsive depopulation of affected herds. Rapid and reliable detection of ASFV infected pigs is critical for the control of ASFV. One desirable property of a diagnostic tests is the capacity to detect viral infection, especially during the incubation time, when the infected animals have not displayed any clinical signs. In this study, we evaluated performance of three pen-side tests for ASFV detection: one PCR test for detection of viral genomic DNA and two lateral flow tests for detection of viral antigens.
The first objective was to determine the time from infection to the earliest detection. Ten pigs were experimentally infected with an ASFV strain. Whole blood and oral swab samples were alternatively collected from five pigs every other day post-infection (dpi) and tested with the three pen-side tests. The pen-side PCR test detected infected pigs starting from 2 dpi when using whole blood and 3 dpi when using oral swabs. It consistently detected infection until the end of the study (10dpi). The antigen test detected infected pigs starting from 3 dpi and no longer detected infection at 10 dpi when using whole blood. The antigen test did not work well when tested with oral swabs. Compared with the reference laboratory real-time PCR test, the pen-side PCR test exhibited 97.8% sensitivity and 100% specificity. The antigen had 100 specificity but only 47.8% sensitivity, mainly because it failed to detect infection from samples collected early or late after infection.
The second objective was to evaluate the diagnostic performance of the tests with samples collected from the field. Whole blood and oral swabs were collected from 205 pigs: 34 positives and 171 negatives as determined by the reference laboratory real-time PCR. All pen-side tests had 100% specificity, regardless of the sample types tested. The sensitivity of the pen-side PCR test was 88.2% and 70.4%, respectively when tested with whole blood and oral swab. The sensitivity of the antigen test 50% and 11.11% respectively when tested with whole blood and oral swabs.
In summary, the results of this study show that the PCR pen-side test has better performance than the antigen test as it can detect infected pigs earlier and for a longer duration after infection than the antigen test. Additionally, the pen-side PCR test works with both whole blood and oral swabs while the antigen test work with only whole blood.
Project #: 21-114 | Principal Investigator: Ganwu Li | Institution: Iowa State University | Posted: 5/25/2022 | Keywords: porcine mobillivirus, virus isolation, real-time RT-PCR, virus isolation, epidemiology
Paramyxoviruses that are known to naturally infect swine include porcine rubulavirus, Menangle virus, Nipah virus, and porcine parainfluenza virus. There are reports of less well-characterized paramyxoviruses associated with central nervous and respiratory disease in pigs. However, none of these viruses are classified in the genus Morbillivirus. In the spring of 2020, the Veterinary Diagnostic Laboratory at Iowa State University received twenty-two porcine fetuses (neonatal mortality, stillbirths, mummified fetuses, and fetuses with moderate autolysis) from six litters submitted from Mexico for diagnostic investigation. PCV2, PCV3, PRRSV, PPV1, and Leptospira sp. were not detected by qPCR or RT-qPCR in any litter. Metagenomics sequencing identified a new virus in the genus of Morbillivirus: Porcine Morbillivirus (PoMV). Other currently known members in the genus Morbillivirus, including measles virus (MeV), rinderpest virus (RPV), peste des petits ruminants virus (PPRV), canine distemper virus (CDV), phocine distemper virus (PDV), cetacean morbillivirus (CMV), and feline morbillivirus (FeMV), are highly contagious pathogens and can cause serious human and animal diseases. Therefore, there is an urgent need to determine if this new virus associated with swine fetal death is present and, if yes, its prevalence in the U.S. swine population. A total of 450 clinical samples (brains, lungs, and spleens from neonatal mortalities, stillbirths, and mummified fetuses) were collected from all over the United States and subjected to PoMV rRT-PCR assay. Our current assessment has not found PoMV in the U.S. swine populations tested thus far. In addition, virus isolation would be valuable for future characterization of PoMV (diagnostics, prevention, pathogenicity, and pathogenesis investigations). Isolation of PoMV in various cell lines (MARC-145 (a clone of the MA-104 cell line, ATCC CRL-2378), MDCK (ATCC NBL-2), PK-15 (ATCC CCL-33), ST (CRL-1746), Vero (ATCC CCL-81), BHK-21 (CCL-10), LLC-MK2 (CCL-7), ZMAC (ATCC PTA-8764), and primary porcine kidney cells using available PoMV PCR-positive tissue homogenates was attempted in this study. Unfortunately, a PoMV isolate that could efficiently replicate in cell culture has not yet been obtained.
Project #: 20-073 | Principal Investigator: Derald Holtkamp, DVM, MS | Institution: Iowa State University College of Veterinary Medicine | Posted: 5/21/2022 | Keywords: PRRSV, PEDV, PDCoV, wean-to-market, biosecurity
The swine industry has committed considerable effort to researching and improving biosecurity practices in swine breeding herds; however, attention to biosecurity in the wean-to-market phase of production lags. Events or characteristics of premises that are more frequently associated with introducing PRRSV and porcine enteric coronaviruses in wean-to-market premises are not well understood compared to the swine breeding herds.
The objective of this study was to detect the introduction of wild-type PEDV, PDCoV, TGEV, and PRRSV into groups of growing pigs and to associate the timing of the introductions with the frequency and timing of pathogen-carrying agent entry events.
Seventy-five groups of growing pigs were enrolled in the study originating from sow farms that were consistently weaning pigs that were negative for PRRSV (AASV Category II-vx or IV) and coronaviruses, including porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV) and transmissible gastroenteritis virus (TGEV). Eight pen-level oral fluid (OF) samples were collected from each group of pigs at placement and every two weeks until the end of the wean-to-market phase for all the groups enrolled in the study and tested via PCR for PRRSV, PEDV, PDCoV, and TGEV. Pathogen-carrying agent entry events and biosecurity characteristics for each group were tracked on a two-week basis for each group of pigs paired with the diagnostic results. The categories included premises demographics, people movement, vehicles/deliveries, carcass disposal, air and water entry, swine movements, manure removal, entry of other animals, and disease status.
Overall, 15.5% of groups became positive for PEDV, 26.4 % became positive for PDCoV, and 97.3% became positive for PRRSV. No cases of TGEV were detected.
During the nursery phase, groups that tested PEDV positive had a higher number of average wean pig delivery events, feed deliveries, propane and fuel deliveries, new supply entries, caretaker visits, and maintenance performed outside the barns than nursery premises that tested negative for PEDV. PDCoV positive nursery groups had a higher average number of wean pig delivery events, feed deliveries, propane and fuel deliveries, new supply entries, transferred supply entries, caretaker visits, maintenance performed inside the barns, maintenance performed outside the barns, visits by management, veterinarians, and other visitors compared to nursery groups that tested negative. Entry of new supplies and maintenance performed inside the barns were more frequent for nursery groups that tested positive for PRRSV than negative groups.
PEDV positive finisher groups had a higher average number of feeder pig delivery events on clean trailers, market pig removals, feed deliveries, propane and fuel deliveries, caretaker visits, maintenance performed outside the barns, and visits by management, veterinarians, and other visitors compared to PEDV negative finisher groups. PDCoV positive finisher groups had a higher average number of feeder pig deliveries, market pig removals, feed deliveries, propane and fuel deliveries, new supply entry, and caretaker visits compared to PDCoV negative finisher groups. PRRSV positive finisher groups had a higher average number of feeder pig deliveries, market pig removals, cull pig removals, feed deliveries, rendering removal, propane and fuel deliveries, transferred supply entry, caretaker visits, and visits by management, veterinarians, and other visitors compared to PRRSV negative finisher groups.
Project #: 21-074 | Principal Investigator: Ganwu Li | Institution: Iowa State University | Posted: 4/30/2022 | Keywords: Streptococcus equi subspecies zooepidemicus (S. zooepidemicus), whole genome sequencing analysis, genomic island, virulence gene, pahtogenicity, epidemiology
High mortality events due to Streptococcus equi subspecies zooepidemicus (Streptococcus zooepidemicus) in swine in the United States were firstly reported in Ohio and Tennessee in September and October 2019. One and a half year later (February, 2021), two-year-old adult sows from a production system in Indiana experienced increased sow death loss with 66 deaths in a 2400 sow on production herd within a six-week period. To investigate if the Indiana outbreak isolates were similar /different to those isolates from Ohio and Tennessee outbreaks, whole genome sequencing analysis was performed. Four outbreak isolates from Indiana were genetically distant to those isolates caused high mortality events in Ohio and Tennessee in the spring of 2019, while closely related to an S. zooepidemicus isolate from a horse in Iowa, suggesting that more than one strain of S. zooepidemicus could cause high mortality events in the United States. The genome sequence of the Indiana outbreak isolate was further closed using Nanopore sequencing, comparative genomic analysis was performed, and genomic islands and putative virulence genes were identified. Two genomic islands (GI-3 and GI-13) were identified specifically only in the Indiana outbreak isolates, thus could serve as the biomarker for the diagnosis of this particular strain. In addition, M-like protein gene (szM) and the Fic domain-containing protein gene (bifA) were positive in those Ohio and Tennessee outbreak isolates, but absent from the Indiana outbreak isolates. Identification of biomarkers of bacterial virulence will significantly improve our response to future possible outbreaks caused by S. zooepidemicus.
View Full Report: As published in peer-reviewed journal October 2023
Project #: 20-178 | Principal Investigator: Cassandra Jones | Institution: Kansas State University | Posted: 1/7/2022 | Keywords: swine, epidemiology, feed safety, porcine delta coronavirus
Two feed mills and three breed-to-wean facilities were investigated after being diagnosed with porcine deltacoronavirus (PDCoV) with initial suspicion that feed manufacture and delivery processes were involved in disease transmission. Both feed mills were audited and environmental samples collected in areas that were deemed high risk for virus contamination. All breed-to-wean facilities had PDCoV detected as would be expected, while the only positive samples for enteric coronaviruses associated with feed mills were feed delivery trucks. These results indicate that feed delivery surfaces can help spread virus during an ongoing disease outbreak and must be considered when determining the outbreak origin.
Project #: 20-172 | Principal Investigator: Cesar A Corzo | Institution: University of Minnesota | Posted: 2/11/2022 | Keywords: monitoring, emerging, PRRS, PEDV, trends
Objective 1 of 4: Monitor trends in pathogens incidence and prevalence – PRRSv, PEDv, PDCoV, Senecavirus and central nervous system associated viruses continue to be monitored. The 2020-2021 season fortunately ended with the fourth lowest PRRSv breeding herd cumulative incidence (25.8%) during the last 11 years of monitoring. During this year we continued to monitor the emergence and dissemination of a new PRRS variant that caused production losses in the Midwest and changed the seasonality pattern historically observed with PRRS with several associated outbreaks occurring during spring 2020. PEDv continued to be present at a low incidence level as the cumulative incidence remained at 3.5%. Our exploratory data analysis showed that reporting PRRS outbreaks and manure pumping activities are associated as 40% of the breaks occurred within 30 days of this event regardless of type of manure storage.
Project #: 21-115 | Principal Investigator: Scott Dee | Institution: Pipestone Research | Posted: /30/2022 | Keywords: swine, feed, soybean meal, extended storage, viral diseases, temperature
Viruses of veterinary significance such as African swine fever virus, foot and mouth disease virus, Pseudorabies virus and Classical swine fever virus are known to survive for extended periods in plant-based feed ingredients imported into North America. To mitigate risk, high risk ingredients, such as oil seed meals, are stored in designated facilities for extended periods under controlled environmental conditions to minimize viral infectivity prior to use. The results from this study suggest that a storage period of 30-days at a temperature of 23.90 C are required to reduce virus infectivity in plant-based feed ingredients such as soybean meal. The outcomes of the study are important, since previous storage periods for feed were based only on mathematical half-life calculations, not controlled studies using live pathogens and representative conditions. We now have for the first time, scientifically sound data based on the use of infectious agents and representative conditions to advise farmers, feed mill operators, federal officials, regulatory and practicing veterinarians, and feed industry leadership on how long and at what temperature to store feed and feed ingredients, to minimize risk. Hopefully, this information will enhance the application and efficacy of Responsible Imports protocols as we collectively work to manage the global risk of feed.
Project #: 20-155 SHIC | Principal Investigator: Chad Paulk | Institution: Kansas State University | Posted: 11/2/2021
Soy-based products are known to pose a viable risk to US swine herds because of their ability to harbor and transmit virus. This project aimed to evaluate soy imports into the US as a whole and from foreign animal disease positive (FAD-positive) countries to determine which products are being imported in the highest quantities and observe potential trends in imports from FAD-positive countries. Import data were accessed through the United States International Trade Commission website (USITC DataWeb) and summarized using R (version 4.0.2, R core team, Vienna, Austria). Twenty-one different Harmonized Tariff Schedule (HTS) codes were queried to determine quantities (metric tonnes, MT) and breakdown of different soy product types being imported into the US from 2015 to 2020. A total of 78 different countries exported soy products to the US in 2019 and 2020 with top contributors being Canada (546,467 MT and 481,497 MT, respectively), India (397,858 MT and 430,621 MT, respectively), and Argentina (122,116 MT and 79,471 MT, respectively). Soy oilcake (582,273 MT) was imported in the largest quantities, followed by organic soybeans (270,194 MT) and soy oil (134,436 MT) for 2020. Of the 78 countries, 46 had cases of FAD reported through the World Organization for Animal Health (OIE) World Animal Health Information Database (WAHIS). Top exporters of soy products to the US from FAD-positive countries in 2019 and 2020 were India (397,858 MT and 430,621 MT, respectively), Argentina (122,116 MT in 2019), and Ukraine (40,293 MT and 56,392 MT, respectively). The risk of FAD introduction to the US through soy imports can fluctuate based on where FAD outbreaks are occurring, shipping methods, and end usage of products. A system to monitor these factors could help make future decisions about trade and risk of FAD introduction to US swine herds. Based on information generated from this project the following frequently asked questions (FAQ) and best practices for importation of soy products were compiled below.
Investigators: Fangfeng Yuan1,2, Xingyu Yan1,2, Brandi Feehan2, Rui Guo2, Yanhua Li2, Giselle Cino2, Ying Fang1, 2*, Douglas Marthaler2*
Institution: 1University of Illinois, 2Kansas State University | Posted: 9/15/2020 | Keywords: porcine sapelovirus, emerging swine virus, PSV monoclonal antibodies, PSV ELISA
An emerging porcine sapelovirus was isolated in a diagnostic specimen from a US swine farm, designated as PSV KS18-01. Full-length genome sequence was obtained through next-generation sequencing. Phylogenetic analysis showed that the virus is more closely related to two Japanese strains but is distantly related to two known US strains. PSV specific diagnostic tools were developed, including the monoclonal antibodies again VP1 and VP2, and a VP1-VP2 antigen-based indirect ELISA. Using this assay, the dynamic response of PSV antibody was investigated in a group of post-weaned pigs that naturally exposed with PSV. The availability of the PSV isolate (KS18-01) and the specific diagnostic reagents and assays provide important tools for PSV control and prevention.
Project #: 20-081 | Investigators: Luis G. Giménez-Lirola, Neeraja Venkateswaran | Institution:Innoceleris LLC. and Tetracore Inc. | Posted: 2/24/2023 | Keywords: Bayesian latent class analysis, ELISA, PCR, African swine fever, diagnostic test, evauation, Vietnam
In the absence of effective African swine fever virus (ASFV) vaccines, infection prevention and control through diagnostic testing and quarantine is critical. Early detection and differential diagnosis of ASFV infections increase the chances for successful control of this devastating disease. However, the interpretation of the ASF diagnostic results can be complicated due to the complex epidemiology of the disease, and its unspecific and highly variable clinical presentation, i.e., same strain producing a wide range of clinical forms. The objective of this proposal was to evaluate the performance of ASFV serum/oral fluid indirect ELISA (iELISA) (collaborative work between Innoceleris LLC. and Tetracore Inc.) for surveillance and monitoring of ASFV outbreaks in commercial farms in Vietnam. For this cross-sectional field study, our field team at Hanoi University collected a 398 paired serum/oral fluid samples, individually collected from each animal, which included 100 samples from 34 ASF-acute farms, 98 samples from 47 ASF-chronic farms, and 200 samples from 20 ASF-negative farms. The samples were tested by Tetracore ASFV iELISA and real-time PCR (qPCR). As expected, the detection rate by qPCR (74% serum; 69% oral fluid) was higher than by ELISA (16% serum; 11% oral fluid) in acute farms due to that most of the animals did not seroconvert yet. Contrary, in chronically affected farms, the detection rate of the ELISA was higher (72% serum; 57% oral fluid) than the qPCR (56% serum; 34% oral fluid). However, when we combined both qPCR and ELISA, the detection rate of ASFV positive animal increased in acute (75% serum; 74% oral fluid) and particularly in chronic farms (85% serum; 74% oral fluid). All serum samples from negative farms were negative by both ELISA and qPCR (100% diagnostic specificity) while, for oral fluids, we obtained 100% and 99% diagnostic specificity for qPCR and ELISA, respectively. The high diagnostic specificity of the tests is particularly important for ASF surveillance. Absence of false positives avoid false alarms and disruption in production, and lack of confidence in the tests/surveillance system. This unprecedented study show that there are no single best diagnostic approach for ASFV surveillance and demonstrate that the combined use of the Tetracore qPCR and indirect ELISA tests and serum/oral fluid sampling, increase efficiency of ASF disease surveillance.
Project #: 20-077 | Investigator: Marie Culhane | Institution: University of Minnesota | Posted: 11/1/2021 | Keywords: African swine fever virus, boar stud, semen, risk assessment
An ASF outbreak in the United States (US) will have a significant impact on the swine and allied industries. This proactive semi-qualitative Risk Assessment (RA) was conducted to evaluate the risk movement of liquid-cooled boar semen (semen) during an African Swine Fever (ASF) outbreak in the US swine industry will result in the spread of ASF virus (ASFv) to other premises with swine. Risks of ASFv spread associated with the movement of semen originating from a Monitored Premises within, into, and outside a Control Area were evaluated as part of a public-private partnership between the US swine industry, the College of Veterinary Medicine at the University of Minnesota (UMN), and the Swine Health Information Center. When completed, reviewed, and cleared, the RA can be used to support permits for the safe and timely movement of fresh boar semen during an ASF outbreak. This assessment is applicable to US swine production and considers the implementation of standard boar stud and semen production practices, enhanced biosecurity recommendations (EBRs) proposed by the Secure Pork Supply (SPS) plan (December 2017), and industry-agreed upon targeted mitigation measures determined by the Boar Stud Working Group (WG).
This RA is limited in scope and intended to identify ASFv transmission pathways associated with semen movement and assess the corresponding likelihoods of carrying ASFv from an infected boar stud premises and causing an infection on off-site sow or gilt breeding facilities during an ASF outbreak in the US, despite the implementation of all standard preventive measures as well as outbreak-specific targeted and/or enhanced measures. This RA will ultimately provide the framework necessary for decision-makers to both quickly assess the effectiveness of preventive measures as they pertain specifically to semen movement and also implement a permit system to allow boar studs with no known infection to move semen into, within, and outside of the ASF outbreak Control Area.
The risk evaluation is based on the likelihood of the presence of infected or contaminated semen products. The likelihoods evaluated include 1. A boar stud facility in a Control Area becoming infected with ASFv; 2. Detecting infected boars on the origin premises prior to semen movement; 3. Semen products becoming contaminated prior to movement off the boar stud; 4. Semen products becoming contaminated in transit by fomites; and 5. ASFv release during transportation of infected or contaminated semen. The likelihood that other susceptible pigs can be exposed to ASFv and become infected as a result of insemination or shipment will be evaluated in cooperation with a swine breeding WG as the proactive RA continues.
The overall findings, which require further evaluation, review, and clearance at the conclusion of the RA process, are that the risk that movement of liquid, cooled boar semen from a boar stud premises that qualifies as a Monitored Premises inside a Control Area to an off-site sow gestation or farrowing facility within or outside of a Control Area during an ASF outbreak results in the infection of susceptible swine at the destination premises is likely to be low to moderate if boar studs follow the Enhanced Biosecurity Recommendations [(EBRs) – as outlined in the Secure Pork Supply (SPS) Plan] and as agreed upon by the WG. In addition, as tactical, targeted biosecurity measures are developed with WG consultation, additional mitigations, meant to be targeted EBRs, and, assuming that these additional, strategic, targeted EBRs will be implemented as mitigation measures, the risk will likely be decreased.
This work is an evolving product-specific risk assessment that will be reviewed and updated as necessary before and during an outbreak to incorporate the latest scientific information, preventive measures, and analytical methodology. Updates and revisions to risk assessments are advisable and necessary as new data become available. The opportunity to update risk assessments to address the limitations of previous analytical methods and data might have effects on the risk ratings and outcomes. If the Incident Command System is activated in response to an ASF outbreak, APHIS (and Incident Command staff) will review this risk assessment with respect to the situation in order to assess industry requests for the movement of boar semen.
Principal Investigators: Encheng Sun, et. al. | Institution: Harbin Veterinary Research Institute | Posted: 10/28/2021 | Keywords: African swine fever virus, geotype I, virulence, pig, China
The Georgia-07-like genotype II African swine fever virus (ASFV) with high virulence has been prevalent in China since 2018. Here, we report that genotype I ASFVs have now also emerged in China. Two non-hemadsorbing genotype I ASFVs, HeN/ZZ-P1/21 and SD/DY-I/21, were isolated from pig farms in Henan and Shandong province, respectively. Phylogenetic analysis of the whole genome sequences suggested that both isolates share high similarity with NH/P68 and OURT88/3, two genotype I ASFVs isolated in Portugal in the last century. Animal challenge testing revealed that SD/DY-I/21 shows low virulence and efficient transmissibility in pigs, and causes mild onset of infection and chronic disease. SD/DY-I/21 was found to cause necrotic skin lesions and joint swelling. The emergence of genotype I ASFVs will present more problems and challenges for the control and prevention of African swine fever in China.
Project #: 20-103 | Principal Investigator: Giovani Trevisan | Institution: Iowa State University | Posted: 10/8/2021 | Keywords: data analysis, surveillance, preparedness, animal health treats, diagnostic data
In 2017 SHIC funded the Domestic Swine Disease Surveillance project, currently housed under the Swine Disease Reporting System (SDRS) initiative. The purpose of this proposal was to keep the Domestic Swine Disease Surveillance program ongoing and further develop a capability to improve sustainability over time. The specific aims included a) keep updated the aggregated database with diagnostic data from the participating laboratories, and provide monthly PDF reports to SHIC as well as keep updated the online interactive dashboards; b) enhance the sustainability of the project by migrating the report-building mechanism to an automated R Markdown technology; c) improve data exchange capability between the Data warehouse project, the Domestic Swine Disease Surveillance program, and other SDRS programs by migrating the data visualization platform to Tableau.
During the time of this project, the database was consistently updated with data from participant laboratories. The database was continuously updated and now contains data from more than 950,000 distinct submissions tested by PCR for the five U.S. porcine endemic agents, i.e., porcine reproductive and respiratory syndrome virus (PRRSV), porcine epidemic diarrhea (PEDV), porcine deltacoronavirus (PDCoV), transmissible gastroenteritis (TGEV), and Mycoplasma hyopneumoniae. Interactive online dashboards with filtering capabilities, e.g., age category, specimen, geographic region, were kept alive, updated, and are available on the project website (https://www.fieldepi.org/sdrs). Monthly PDF reports have been consistently produced and releases every first Tuesday of the month. Monthly PDF reports are published at the project webpage, SHIC website (https://www.swinehealth.org/domestic-disease-surveillance-reports/), and distributed by email to more than 270 registered receivers from 129 organizations/institutions from 7 countries (US, Canada, Mexico, Brazil, Chile, Germany, and Spain).
In July 2020 (SDRS report # 29), monthly PDF reports started to be produced using the R Markdown technology and gained a new face. An R Markdown script has written and allowed the generation of reports in a standard and consistent format. The R Markdown script was codified to automate as much as possible the process of generating and update images, formatting, and description for the monthly detection changes under each agent page. As an ongoing real-time project, there is still the need for staff to interpret the finding, debugging, communicate results with the advisory group, gather feedback, and compile the final report, including the input and feedback from the Advisory group. The usage of an R Markdown script has enhanced SDRS sustainability by reducing the time needed and errors in compiling the final monthly PDF final report.
Interconnection between databases and different visualization and analytical tools are key elements for project sustainability. The need to efficiently process and interconnect the database with the R Markdown and visualization platforms like Power BI and Tableau requested a redesign of the SDRS database structure. The SDRS has been built using a building block approach. Initially, SDRS started reporting PRRSV detection from Iowa and Minnesota VDLs and later on added South Dakota and Kansas VDLs and additional data for the enteric coronavirus and M. hyopneumoniae. Initially, each agent’s database was kept separated. A restructuring was done to compile, combine, and store all data into a single database. This redesign allowed the database interconnection with the R Markdown to generate PDF monthly reports, analytical tools like R and SAS, and business intelligence tools, like Power BI and Tableau, for data visualization.
Additionally to the proposed scope of this project, the SDRS team has consistently produced monthly audio and video reports distributed on the project website, Youtube Channel, Swine Cast platform, and LinkedIn. Also, during the last year, SDRS has been actively engaged in providing information and support to the U.S. swine industry when it went into a crisis due to the activity of contemporary and emergence of a new PRRSV strain threatening the U.S. swine health. Early detection is a key component for containing the spread of emerging or re-emerging animal health threats. More than 110 monitoring algorithms are currently implemented in the SDRS background to detect early changes in the pattern of agent detection.
This collaborative project funded by SHIC has real-time capabilities for data collection, analysis, and sharing of swine health data and information to protect and enhance the health status of the United States swine herd. SDRS is a real-time coordinated and largely representative national swine disease monitoring program that provides VDL information in efforts to minimize disease threats current and future impact. The SDRS is the only publicly available source of swine health information from U.S. VDLs. It is also the only source of information on pathogen activity in all age groups (from boar studs to grow-finish pigs). The sharing of information on endemic and endemic re-emerging diseases affecting the swine population in the U.S. has and continues to assist veterinarians and producers in making informed decisions on disease prevention, detection, and management.
Project #: 21-065 | Principal Investigator: Diego G. Diel | Institution: Cornell University | Posted: 10/5/2021 | Keywords: feed, biosecurity, nucleic acid extraction, PRRSV, SVA, PEDV
Feed biosecurity has been an area of significant interest to the swine industry. Early studies with porcine epidemic diarrhea virus (PEDV) suggested that the virus may have been transmitted through feed. Recent experimental evidence confirmed that PEDV, SVA and FMDV can indeed be transmitted through contaminated feed that is ingested naturally by susceptible pigs. One way of reducing the risk of pathogen transmission through feed is to test feed ingredients and feed before they are introduced into farms and fed to pigs. This would only be possible if sampling and nucleic acid extraction methods would allow efficient detection of pathogens in feed. In this study we focused on comparing the performance of three commercially available nucleic acid extraction kits (CORE, IndiMag, MVP II). These kits were tested in samples that were spiked with PRRSV, SVA and PEDV and that were previously collected as part of a transportation study and tested in another VDL. Our results show that the Core extraction kit outperformed the other two kits evaluated in the present study and previously used in another VDL that originally had tested the samples. Overall samples extracted with the Core kit presented lower Ct value (at least for PRRSV and SVA) and a higher sensitivity when compared to samples extracted with MVPII or the IndiMag, One of the key issues that remains to be addressed in future studies is the sampling method to be used for large volumes of feed or feed ingredients.
Project #’s: 18-137 and 18-211 | Investigators: Leonardo C. Caserta1,2, Jessica C. G. Noll1,2,Aaron Singrey1,Megan C. Niederwerder3,Scott Dee4,Eric A. Nelson1,Diego G. Diel1,2 | Institution: 1South Dakota State University, 2Cornell University, 3Kansas State University, 4Pipestone | Posted: 9/1/2021 | Keywords: feed biosecurity, half-life, ingredients, Senecavirus A, SVA, swine diseases
Animal feed and feed ingredients have recently been investigated as sources of pathogen introduction to farms and as a potential source of infection to animals postconsumption of contaminated feed. Survival of several viruses for a prolonged period has been demonstrated in feed. Here, we determined the rate of decay of Senecavirus A (SVA) in swine feed ingredients as a function of time and temperature and established half-life estimates for the virus. Select feed ingredients were spiked with a constant amount of SVA (105 median tissue culture infectious dose 50) and incubated at 4, 15 and 30◦C for up to 91 days. Virus viability and the presence of viral RNA were assessed in samples collected over time. At the three different temperatures investigated, dried distillers’ grains with solubles (DDGS) and soybean meal (SBM) provided the most stable matrices for SVA, resulting in half-lives of 25.6 and 9.8 days, respectively. At 30◦C, SVA was completely inactivated in all feed ingredients and in the control sample, which did not contain a feed matrix. Although virus infectivity was lost, viral RNA remained stable and at consistent levels throughout the experimental period. Additionally, the ability of SVA to infect swine via ingestion of contaminated feed was investigated in 3-week-old, weaned pigs. Animals were provided complete feed spiked with three concentrations of SVA (105, 106 and 107 per 200 g of feed) and allowed to naturally consume the contaminated feed. This procedure was repeated for three consecutive days. Infection of pigs through consumption of contaminated feed was confirmed by virus neutralization assay and the detection of SVA in serum, feces and in the tonsil of exposed animals by real-time reverse transcriptase PCR. Our findings demonstrate that feed matrices are able to extend the survival of SVA, protecting the virus from decay. Additionally, we demonstrated that consumption of contaminated feed can lead to productive SVA infection.
Project #: 18-194 | Investigators: Carolina Stenfeldt1,2, Miranda Bertram1, Haillie Meek1,3, Ethan Hartwig1, George Smoliga1, Megan Niederwerder2, Diego Diel4, Scott Dee5, Jonathan Arzt1 | Institution: 1Plum Island Animal Disease Center, 2Kansas State University, 3Oak Ridge Institute for Science and Education, 4Cornell University, 5Pipestone | Posted: 7/6/2021 | Keywords: contamination, feed, foot-and-mouth disease, foot-and-mouth disease virus, pig
Important work examining the role of contaminated feed as a vector for transmission of foot-and-mouth disease virus (FMDV)was published in July 2021. Specifically, the study performed by researchers from ARS/USDA at Plum Island, evaluated the potential risk of incursion of FMDV into naïve pig herds through contamination of feed. Per the study report, the goal of the project was the assessment of the infectiousness (viability) of FMDV in commercial whole pig feed and pig feed ingredients. Additionally, the researchers, led by Drs. Stenfeldt and Arzt, determined the dose required to infect pigs through natural feeding behavior. Finally, the project looked at the ability of select commercially available feed additives to reduce infectivity of contaminated feed. “While comparable research investigating the potential biosecurity risks of imported feed exists for other viral pig pathogens (Dee et al., 2018; Niederwerder et al., 2020; Niederwerder et al., 2019), this is the first comprehensive evaluation of the risk of FMDV infection of pigs through ingestion of contaminated feed under controlled experimental conditions,” wrote the authors.
Project #: 18-142 | Principal Investigator: Christa Goodell | Institutions: Boehringer Ingelheim Vetmedica GmbH, Ingelheim | Posted: 6/2021
Vietnam has lost ~6 million pigs to ASFV since the first reported outbreak in February 2019. The national swine herd has returned to 88.7% of its pre-ASFV level, but the risk of ASFV recurrence is high because of new and on-going ASFV outbreaks (Report #VM2021-0042, USDA & GAIN, May 11, 2021).
Because of the increased value of pork in Vietnam due to reduced supply, standard ASFV control measures have centered around a modified “test-and-remove” or “Tooth Extraction” protocol. A common “Tooth Extraction” protocol for a sow farm is to remove any sow exhibiting clinical signs compatible with ASF (taking a whole blood sample for ASFV PCR testing) plus the two sows in stalls on each side of the index (clinical) animal (Yaros, Leman, 2019).
The objective of this study was to test the efficacy of the “Tooth Extraction” protocol.
For each “ASFV event” (identified by farm caregiver), whole blood samples were collected from the index sow plus 14 animals in gestation stalls on each side of the index sow (see Figure 1). Samples were tested for ASFV DNA by real-time ASFV PCR within 24-hours of arrival to the laboratory. The proportion of positive animals was analyzed as a function of the gestation stall distance from the index sow.
Figure 1. Blood sampling protocol around a suspected ASF clinical sow in gestation†‡
†F0=index sow, F1= direct/closest contact neighbors, F2= indirect contact neighbors; A= ”down” row, B= “up” row
‡(The rationale for the sampling distribution was due to an assumption drinking water flowed down a common water trough)
Results. 766 whole blood samples from 52 ASFV events were collected and tested for ASFV DNA by PCR. 85 samples were positive for ASFV DNA by PCR.
Key learnings:
• In 17 (33%) of the 52 events, the index sow and 14 neighbor sows were ASFV PCR negative.
• In 35 (67%) of the 52 events, the index sow was ASFV PCR positive.
– Among allsows detected as ASFV PCR positive, 39 (78%) were located outside of the index animal and her direct contact neighbors (F0, F1A1, F1B1). Thus outside of the index animal and her contact neighbors (F0, F1A1, F1B1), 10% of all animals tested were ASFV PCR positive, compared to the direct contact animals alone (F1A1, F1B1), representing 18% of the all animals tested.
– If the index sow and 4 direct/closest contact neighbors were removed (F0, F1A1, F1B1, F2A1, F2B1), there was a 50% probability that additional ASFV PCR positive (but unidentified) sows remained.
• ASFV DNA was detected in blood from sows showing no clinical signs.
The results of this study suggest “Tooth Extraction” is not sufficient to eliminate ASF from a pig farm.
Project #: 20-175
Discovered in 2001, porcine parvovirus 2 (PPV2) is prevalent in swine in countries worldwide. While it is commonly detected in swine samples, the clinical significance of infection is unclear. We recently identified PPV2 present at high levels in the lung of a pig with pneumonia, prompting this investigation into the role of PPV2 in respiratory disease. Here we report that PPV2 was detected in 39% of lungs submitted for routine diagnostic testing. There was a significant positive correlation between PPV2 viral load and pig age. Histologically, PPV2 was mainly detected in alveolar macrophages in 28% of lungs with interstitial pneumonia, with PPV2 viral load tending to correlate with the number of macrophages in the lungs. While co-infections with PPV2 and established swine respiratory pathogens influenza A virus and porcine reproductive and respiratory syndrome virus were commonly detected, assessment of viral loading and tissue locations showed no obvious association between PPV2 and other viral pathogens. Relatedly, in a third of PPV2 positive lungs analyzed by metagenomic sequencing, no other pathogens were identified. Together, these results suggest that PPV2 may play a primary role in porcine respiratory disease.
Project #: 19-154 | Principal Investigator: Daniel C. L. Linhares, DVM, MBA, PhD | Institution: Iowa State University | Posted: 2/9/2021 | Keywords: biosecurity, vulnerability, scoring, risk, swine, breeding herd, PRRS
PRRS continues to plague the swine industry with 20-30% of herds breaking annually. These outbreaks cost the swine industry and its producers $664 million every year. Swine producers continue to offer efforts to increase biosecurity with the intention to provide the best welfare for their pigs and produce a wholesome product for consumers. The industry needs the ability to better predict risk of a PRRS outbreak as well as to evaluate the level of biosecurity in their farms. The objective of this study was to measure and benchmark the relationship between key biosecurity aspects and PRRS outbreaks in breeding herds, while validating a short biosecurity screening survey (44 questions). The survey captured data on herd demographics, PRRS outbreak history, frequency of high-risk events, surrounding swine density, transport biosecurity practices, carcass disposal, and people movement. Three machine-learning algorithms were used to identify key biosecurity factors and practices associated with PRRS outbreaks. The most accurate, based on ability to explain variability between farms in the frequency of reported PRRS outbreaks, was selected. Moreover, positive predictive value (PPV) from the models were utilized as a biosecurity score. We investigated the correlation between PPV and the reported frequency of PRRS outbreaks.
Thirteen production systems from 15 states were enrolled in the study (n=188 sow herds). The best machine learning algorithm predicted PRRS occurrences with an accuracy of 78.5%. The 44 biosecurity measures were ranked according to their contribution to the model prediction. The first four most important variables were devotion to breeding genetic replacements, number of swine premises within a three-mile radius, number of breeding females on the premises, and distance to the nearest public road. The probability that a herd had reported an outbreak were higher in farms that had raised breeding animals as genetic replacements. The higher the number of swine facilities within a three-mile radius and the closer the farm was to a public road, the more likely the facility would be expected to break with PRRS. Finally, the PPVs were correlated with the number of reported PRRS outbreaks (correlation of 72%). The majority of farms that had reported no outbreaks in the past five years had low (<50%) probability of having a PRRS outbreak (i.e., low score) while a larger number of PRRS cases were expected for farm who had reported at least one outbreak (i.e., high score), with few exceptions.
The analysis offers a flexible, shortened approach to screen breeding herd’s PRRS biosecurity vulnerability. This study highlights the value of using data to build upon our understanding of biosecurity risk in an operation.
Project #: 20-071 | Principal Investigator: Hiep Vu| Institution: University of Nebraska-Lincoln | Posted: 2/1/2021 | Keywords: ASFV, heat treatment, swine feces, disinfection, virus inactivation
African swine fever virus (ASFV) is the most devastating viral pathogen which can cause up to 100% mortality in domestic pigs. Currently, the virus is affecting many swine producing countries, leading to the approximately 25% reduction of global swine population. Swine transportation plays a major role in spreading infectious pathogens. Thus, developing a procedure for effectively disinfecting animal trailers will help reduce viral spreading. The most effective method to effectively inactivate pathogens in transportation trails is a combination of washing, disinfecting and drying. However, there are not enough washing facilities to wash all trailers between loads of swine. Therefore, we are interested in testing if it is possible to effectively inactivate ASFV in the presence of organic materials (feces, bedding) through the use of thermal-assisted drying and decontamination (TADD) which commonly operates at the temperature between 63°C and 71°C (Dee et al., 2005).
The objective of this project is to determine the optimal baking time and temperature required to completely inactivate ASFV in aluminum surface contaminated swine feces. We tested the inactivation efficiency of contaminated trays under 3 conditions:
• Baking contaminated aluminum trays at 54oC and 63oC for 5, 10, and 15 min
• Power washing the tray surface with water at room temperature followed by baking at 54oC and 63oC for 5, 10, and 15 min
• Power washing the tray surface with water, spray disinfectant (Virkon-S) followed by baking at 54oC and 63oC for 5, 10, and 15 min
Two different methods were used to evaluate the efficiency of the treatments: PCR to detect of viral genomic DNA and virus isolation to detect infectious virus.
In the first condition, swabs collected from contaminated trays at all time-points post incubation at 54oC and 63oC were positive by PCR, indicating that heat treatment could not eliminate viral genomic DNA. On the other hand, swabs collected from contaminated tray at 5 min post incubation at either 54oC or 63oC were negative by virus isolation, indicate that holding ASFV in the presence of feces at 54oC for 5 min is sufficient to inactivate the virus.
In the second and third conditions, only two swabs collected after washing were positive by PCR at high Ct value (e.g. 37.03). These swabs were negative by for virus isolation. Thus, under the conditions of this study, power washing of the trays with water at room temperature was efficiently remove contaminated material from the trays.
Collectively, results obtained from this research provide valuate information for the develop effective sanitation protocols to disinfect animal trailers to reduce the spreading of ASFV.
Project #: 19-236 | Principal Investigator: Nubia Macedo | Institution: Iowa State University | Posted: 1/31/2021 | Keywords: Streptococcus equi subsp. zooepidemicus, swine, experimental model, PCR
In swine, Streptococcus equi subsp. zooepidemicus (SPZ) caused a severe outbreak in China in 1975. In the United States, a genetically similar and hypervirulent SPZ strain caused high mortality outbreaks in sows and feeder pigs in 2019. There is currently no available challenge model in pigs to evaluate the disease process and diagnostics, which would provide meaningful information about the pathogenicity and epidemiology of SPZ in pigs. To address this need, conventional 6-week-old pigs were challenged using hypervirulent SPZ strains and a genetically different, less virulent SPZ strain. Additionally, sequenced SPZ strains were assessed to identify specific virulence markers, and a multiplex RT-PCR assay was designed for SPZ identification and prediction of virulence in swine isolates. Pigs challenged with the hypervirulent SPZ strains developed severe systemic disease, with mortality varying from 50-100%. In contrast, pigs challenged with the less virulent swine strain had minimal clinical signs and no mortality. The colonization and systemic distribution of SPZ were evident in challenged pigs by culture and PCR. This is the first study to experimentally infect and reproduce the disease in weaned pigs with a hypervirulent swine SPZ strain. Furthermore, pathogenicity differences between genetically different swine strains were described. This experimental model will allow for further investigations on the pathogenesis of SPZ in swine and the development of methods of prevention and control of this emerging disease. The newly developed multiplex RT-PCR provides an accurate and timely assay for detecting and monitoring SPZ in swine herds.
Project #: 19-235 | Principal Investigator: Cesar A Corz | Institution: University of Minnesota | Posted: 1/20/2021 | Keywords: monitoring, emerging, PRRS, PEDV, trends
Objective 1: Monitor trends in pathogens incidence and prevalence – PRRSv, PEDv, PDCoV, Senecavirus and central nervous system associated viruses continue to be monitored. The 2019-2020 season fortunately ended with the third lowest PRRSv breeding herd cumulative incidence (24.5%) during the last 11 years of monitoring. During this year we saw a different trend in breeding herd PRRSv prevalence as the proportion of herd staying in category 1 increased and “plateaued” which had not been seen before. PEDv and PDCoV continued to be present at a low incidence level. Mycoplasma hyopneumoniae was included into our monitoring program through a convenient sample of 8 systems. We now have the ability to quickly add new pathogens, which allow us to be prepared in the case of a FAD introduction.
Objective 2: To conduct prospective monitoring of PRRSv sequence evolution and impact – PRRSv sequence acquisition has been stabilized and these are being acquired on a monthly basis. Throughout the year, several participants contacted us to provide outbreak investigation support. A total of 29 sequence analysis were conducted with one of the latest being a virulent 1-4-4 virus. This allowed us to become a neutral third-party curating sequence to identify similarities and putting systems in contact whenever both parties agree.
Objective 3: Develop capacity to capture and analyze movement data – Transport data is acquired actively and has been analyzed. Movement data can be obtained at a granular level allowing for traceability but most importantly, allows the producer to follow the truck in real-time. Transport biosecurity compliance continues to become an achievable goal including every single step between the truck-wash, loading of pigs, unloading and return to truck-wash. Characterization and description of transport data has shown that few transport vehicles come in contact with 1/3 of the farms of the participating production system highlighting an important level of connectivity.
Objective 4: To expand participation of producers to allow for all to be involved – Expansion continues at three levels: sow, boar and growing pig populations. A new production system joined the SHMP during the year. A total of 30 boar studs from 12 participants have been added to our database. The growing pig population continues to grow with 4 companies sharing their growing pig locations. Work is being done towards linking sow-growing pig populations in our database.
Project #: 19-229 | Principal Investigator: Andres Perez | Institution: University of Minnesota | Posted: 11/2020 | Keywords: swine diseases, global surveillance, hazards preparedness, awareness
The expansion of ASF through Asia has raised concerns in the industry, resonating the events of 2013, when a devastating epidemic of porcine epidemic diarrhea (PED) virus caused far-reaching losses. Those events demonstrated the importance of developing systems to provide situational awareness to stakeholders in near-real time, to coordinate actions between government agencies and the industry with the ultimate objective of preventing or at least mitigating the impact of diseases epidemics. The swine industry is vulnerable to the introduction of pathogens, and their variants, from which the US is currently free. We have developed a private- public-academic partnership to support a system for near real time identification of hazards that will contribute to the mission of assessing risks to the industry. Identified hazards were shared monthly with swine practitioners and the government, to help increase the country awareness and preparedness. Ultimately, the system has kept contributing to identify and early detect or create awareness on key stakeholders to support current prevention and mitigation strategies for introduction of foreign pathogens into the US.
Project #: 19-153 | Principal Investigator: Kimberly VanderWaal | Institution: University of Minnesota | Posted: 11/1/2020 | Keywords: prediction, disease, forecast, infection, swine, virus, PEDV, epidemiology
Despite our growing understanding of the epidemiology of viral diseases such as Porcine Epidemic Diarrhea virus (PEDv) and Porcine Reproductive and Respiratory Syndrome virus (PRRSv), a gap remains between the science and the ability of producers to be able to effectively estimate and respond to spatial and temporal variation in risk. Therefore, the aim of this project was to generate farm-level forecasts of PEDv risk that account for recent animal movements, present disease distribution, and environmental factors. Utilizing data captured by the Morrison Swine Health Monitoring Project, we built machine learning algorithms that predict whether a sow farm will break with PED two weeks in advance.
Our forecasting tool was able to detect approximately 20% of the outbreaks in a region (sensitivity), while 70% of the outbreaks our tool said would occur did indeed happen (positive predictive value).The most important factors related to breaks were animal movements both into the sow farm and its neighbors, as well as the PED-status of the origin farm of these movements. Farm density and ambient temperature were also important predictors. We developed this tool using data for a single US swine-dense region and began applying it to a second region of the country, operating with different industry systems and a different environment. This forecasting tool is operationalized for real-time use – since December 2019, partnering systems have received weekly system-specific predictions of PEDv outbreak risk at farm level. These predictions are made two weeks into the future, which allows systems, veterinarians and producers to act in case a high probability of an outbreak is forecasted. This provides an important tool for informed decision-making and coordinated actions of producers and practitioners to control or mitigate the impact of PEDv outbreaks.
Project #: 19-237 | Principal Investigator: Derald Holtkamp | Institutions: Iowa State University | Posted: 10/2020
UVC germicidal chambers are used in swine settings to reduce the microbial load on surface items. Chambers, which may be commercial or homemade, are usually constructed so items to be disinfected are passed through from the dirty side (entry/hallway) to the clean side (office/break room).
UVC germicidal chambers are mostly used for small to medium items like lunch boxes, cell phones, small tools, and medications. Food and semen bags can also be passed through the chamber without negative effects. Repeat exposure of plastics to UVC light may lead to a change in the color or smell of the object. Paper and cardboard cannot be disinfected in a UVC germicidal chamber. Larger UVC chambers, or UVC rooms, can be built for larger items.
Project #: 19-211 | Principal Investigator: Gustavo Machado
Institutions: North Carolina State University | Posted: 10/5/2020 | Keywords: swine disease spread, transmission dynamics, disease surveillance, mechanistic modeling
A limited understanding of transmission dynamics is a major obstacle to prevent and control disease spread among swine populations. Forecasting outbreaks before they occur would allow the swine industry to tailor control strategies and improve pig production. Understanding what combination of strategies may be required to reduce between-farm transmission is key to maintain control of outbreaks while minimizing disruptions to pig production. Our objective is to forecast weekly Porcine Epidemic Diarrhea virus (PEDV) outbreaks by generating high-resolution maps to identify current and future PEDV high risk areas, and simulating the impact of control measures. Three epidemiological transmission models were developed and compared: a novel epidemiological modelling framework was developed specifically to model swine populations, PigSpread and two models built on previously developed ecosystems; SimInf (an stochastic disease spread simulations) and PoPS (Pest or Pathogen Spread). Prediction accuracy across models was compared using receiver operating characteristic (ROC) area under the curve (AUC). The models were calibrated on true weekly PEDV outbreaks from three spatial related swine companies. Model outputs had general agreement with observed outbreaks throughout the study period. PigSpread had an AUC of 0.71, SimInf had an AUC of 0.59 and PoPS had an AUC of 0.80. Our analysis estimates that the combined strategies of herd closure, feedback and reinforcement of on-farm biosecurity reduce 14% the incidence of outbreaks in sow and 20% in Gilt Development Units (GDU) when deployed weekly in sow and GDU farms located in risk areas. Accurate forecasts of PEDV spread are feasible to be generated within a decision making timeframe, but the predictability across all models depends on the stage of the epidemic.
Project #: 19-220 | Principal Investigator: Ganwu Li |
Institutions: Iowa State University | Posted: 2/10/2021 | Keywords: diarrhea, phylogenetic analysis, porcine sapovirus, real-time PCR, recombination
A case from the farm experiencing an ongoing problem with piglet diarrhea in the lactation phase for more than 2 years. The pigs exhibited a self-limiting diarrhea starting around 10 days of age, but typically lost 1-2 lbs of expected weaning weight. Histopathological examination of five clinically affected piglets revealed that 5/5 small intestines had moderate villus atrophy, vascular congestion, and lymphocytic infiltration in the small intestine suggestive of an enteric viral infection. Porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus, transmissible gastroenteritis virus (TGEV), or rotavirus were not detected from small intestines using real-time PCR assays (rRT-PCR). Additionally, there was no significant bacterial growth from the small intestines. Herein, we document the discovery of porcine sapovirus of genogroup III as the cause of the enteritis and diarrhea use of four independent lines of evidence: metagenomics analysis, real-time RT-PCR, histopathology, and in situ hybridization. To our best knowledge, this is the first evidence that SaV likely serves as the sole etiological agent causing enteritis and diarrhea of piglets in the field in the United States. In addition, a highly sensitive and specific real-time RT-PCR for detecting porcine sapovirus of genogroup III was established. Prevalence survey of more than 500 samples from both pigs with clinical diarrhea and clinical healthy pigs suggests that porcine sapovirus III play an important role in causing swine enteritis and diarrhea and rRT-PCR is a reliable method to evaluate the pathogenicity role of porcine SaV.
Project #: 16-250 | Principal Investigator: Orlando Perez | Institutions: National Centre for Foreign Animal Disease, Canadian Food Inspection Agency | Posted: 8/31/2020 | Keywords: pseudorabies virus, variant, highly pathogenic, triplex PCR, detection, differentiation, DIVA
Pseudorabies virus (PRV) causes Aujeszky’s disease or pseudorabies (PR), fatal encephalitis in newborn piglets, respiratory infection in growing and fattening pigs, and reproductive failures in pregnant sows. It establishes a lifelong latent infection in the peripheral nervous system followed by subsequent intermittent shedding of infectious virus. Since 2011, highly virulent PRV strains that are genetically different from the classic PRV strains surfaced in pig herds in China. Availability of a highly sensitive and specific polymerase chain reaction (PCR)-based diagnostic assay for rapid differential detection of PRV variants is critical to prevent huge economic losses to the U.S. and Canadian pork industries if these strains enter North America and cause an outbreak. Here we describe the development and evaluation of a single-tube triplex real-time-PCR assay for differential detection of variant strains of PRV. The assay targets the intergenic region between the US2 and US6 genes in the PRV genome and is highly sensitive and specific and it did not detect other nontarget viruses including related herpesviruses. The clinical specificity and sensitivity of the assay was evaluated using whole blood, serum, tissue and swab samples collected from known negative and experimentally inoculated pigs with either classical (Bristol) or variant (JS-2012 and HeN1) PRV strains. The targeted genomic region of this assay is also deleted in commonly used PRV gE-deleted marker vaccines, and therefore, the triplex assay did not detect viral DNA extracted from two commercial vaccine strains Bartha K-61 and Bucharest. This single-tube triplex assay can be used for routine diagnostics and epidemiological studies for detection and differentiation of classical strains from variant strains of PRV, and as a differentiation of infected and vaccinated animals (DIVA) assay when PRV gE- deletion mutant marker vaccines are used.
Project #: 19-149 |Principal Investigator: Derald Holtkamp, PhD | Institution: Iowa State University | Posted: 8/21/2020 | Keywords: epidemiological outbreak investigations, rapid response corps, Rapid Response to Emerging Disease Program
The need to quickly identify, control, and eliminate a pathogen in an endemic, emerging, or transboundary disease outbreak in the United States is crucial to protect the swine industry from suffering huge economic losses. In August of 2016, SHIC funded Iowa State University to develop the Rapid Response to Emerging Disease Program (RRP). A Rapid Response Corp (RRC) was formed for the program. The RRC is a nationwide network of veterinarians, state animal health officials or representatives, epidemiologists and, when appropriate, federal animal health officials who are trained, prepared and committed to moving within 24 hours of contact to conduct epidemiological investigations when a new transboundary or emerging disease threat occurs. The standardized approach, methodology, forms, and reports developed for the Iowa Pork Producers Association (IPPA) funded PRRS outbreak investigation pilot project, were adapted to develop the RRP. Online training modules, checklists, investigation forms, surveys, and report templates to conduct a rapid response investigation are now available for download on the SHIC website. The objective of this project was to facilitate the continuation of the RRP beyond the current five-year funding stream for SHIC by automating and streamlining the rapid response investigation process through a web application. This proposal included the planning phase to automate and streamline the process. The RRC was maintained at 35 members for the duration of this project. To support the continuation of the RRP, and to provide members of the RRC with additional training, a pre-conference seminar was conducted at the American Association of Swine Veterinarian’s Annual Meeting in March of 2020. The seminar was titled “Conducting effective outbreak investigations: Learning from our mistakes, part 2.” The seminar followed the successful pre-conference workshop offered as training for the RRC members at the same meeting in 2019. The possibility of integrating the RRP into the National Pork Board’s swine business continuity system (AgView®) was explored. Delays in the development of AgView led to the exploration of simpler, more executable approaches. A web-based version of the investigation form used by the RRC members to conduct outbreak investigations has been developed using Qualtrix, which is a commercially available online survey platform. The web-based version of the investigation form will be tested in Vietnam, as part of another SHIC funded study to investigate African swine fever (ASF) virus on farms in Vietnam.
Project #: 19-148 |Principal Investigator: Cassandra Jones | Institution: Kansas State University | Posted: 2/25/2020 | Keywords: porcine, Senecavirus, virus, feed, ingredient, sampling, biosecurity
Environmental monitoring is commonly used in pharmaceutical, human food, and pet food manufacturing facilities manufacturing as an indicator of pathogenic bacteria in the product. A correlation between the presence of Salmonella spp. and Enterobacteriaceae within feed mills has been demonstrated, but little information is available on how the presence of Enterobacteriaceae (EBAC) correlates with viral pathogen presence, especially on farms or in feed mills. The purpose of this study was to identify Enterobacteriaceae presence in the feed manufacturing facilities of a multi-farm system experiencing a viral outbreak as a method of identifying biosecurity gaps.
Project #: 19-219 | Principal Investigators: Derald Holtkamp, DVM, MS | Institution: Iowa State University College of Veterinary Medicine | Posted: 12/2019
The swine industry has focused much of its efforts on improving biosecurity in breeding herds; while little attention had been paid in wean-to-finish growing sites. One risk event that has the potential to introduce virus into grow-finish pigs is load-out during marketing. In order for the remaining pigs in the group to become infected during load-out, viral contamination must be transferred from the contaminated livestock trailer, driver or other carrying agents to the pigs in the barn. Unfortunately, little research has been done to assess how frequently this occurs or to assess alternative biosecurity measures to reduce the frequency.
Project #: 19-236 | Principal Investigators: Nubia Resende-De-Macedo | Institution: Iowa State University | Posted: 3/24/2020 | Keywords: bacterial pathogens, emerging diseases, veterinary epidemiology
High mortality events due to Streptococcus equi subspecies zooepidemicus (Streptococcus zooepidemicus) in swine have not previously been reported in the United States. In September and October 2019, outbreaks with swine mortality up to 50% due to S. zooepidemicus septicaemia were reported in Ohio and Tennessee. Genomic epidemiological analysis revealed that the eight outbreak isolates were clustered together with ATCC 35246, a Chinese strain caused outbreaks with high mortality, also closely related to three isolates from human cases from Virginia, but significantly different from an outbreak-unrelated swine isolate from Arizona and most isolates from other animal species. Comparative genomic analysis on two outbreak isolates and another outbreak-unrelated isolate identified several genomic islands and virulence genes specifically in the outbreak isolates only, which are likely associated with the high mortality observed in the swine population. These findings have implications for understanding, tracking and possibly preventing diseases caused by S. zooepidemicus in swine.PCV3 may cause death in fetuses and myocarditis and systemic vasculitis in pigs.
Project #: 18-201 | Principal Investigator: Albert Rovira | Institution: University of Minnesota | Posted: 3/30/2023 | Keywords: PCV3, abortion, myocarditis, vasculitis, diagnostic laboratory, lesions, clinical signs
Porcine circovirus type 3 was discovered in 2016 in the US associated with cases of systemic disease and reproductive disorders. Multiple studies performed during the last two years have shown that this virus is widespread and has been around for decades. It can be found in multiple tissues and samples, in pigs with multiple clinical conditions and in healthy pigs. However, we are lacking precise information on the relevance of this virus and its potential to cause disease in pigs. The objective of this proposal was to mine the diagnostic data obtained by the MN VDL during the last 2 years to identify associations between the presence of PCV3 and its viral load and specific lesions and clinical conditions. PCV3 results, clinical signs and information on the lesions for each pig were retrieved from the MN VDL laboratory information management system, submission forms and diagnostic reports. PCV3 frequency in pigs with different clinical signs ranged from 12% to 27%. No significant associations were observed between clinical signs and the presence of PCV3. In PCV3-positive pigs, no clinical signs were significantly associated with having a higher load of PCV3. PCV3 frequency in pigs with different lesions ranged from 0% to 62%. The only lesion that had a significant association between its presence and PCV3 infection was heart vasculitis/perivascultis. In PCV3-positive pigs, higher viral loads were significantly associated with pigs with myocarditis, heart vasculitis/ perivasculitis, kidney vasculitis/perivasculitis and dermatitis. This study did not identify any significant associations with clinical signs. However, the presence of PCV3 in 20% of fetuses is remarkable. In addition, the samples with the highest PCV3 concentration in this study were from fetal tissues. Lesions of myocarditis and systemic vasculitis were associated with the presence or the amount of PCV3 in tissues. The lack of significant results for other lesions does not exclude the possibility of a real association and may be due to confounding factors or limited data. In summary, this study provides an objective view of the relationship between PCV3 and disease, based on a large dataset of diagnostic cases. PCV3 is a very common virus that circulates in healthy populations and can be detected in around 20% of the pigs submitted to the diagnostic laboratory. Therefore, it is important to differentiate when PCV3 plays a significant role and when it does not. The results from this study support previous studies that suggested that PCV3 may cause death in fetuses and myocarditis and systemic vasculitis in pigs.
Project #: 20-072 | Principal Investigator: Scott Dee | Institution: Pipestone Applied Research | Posted: 5/11/2020 | Keywords: animal feed, feed mitigation, ice-block challenge, swine viral diseases
In 2014, contaminated feed was identified as a vehicle for the transport and transmission of PEDV. Over time, similar conclusions were drawn regarding the role of feed and the risk of African swine fever virus, Senecavirus A (SVA), Pseudorabies virus and Classical swine fever virus. However, as these results were based on laboratory studies, our goal was to validate these observations using a real-world “demonstration project” approach, simulating the actual transport of feed contaminated with SVA, PEDV and PRRSV across the United States in a commercial transport vehicle. Samples of conventional soybean meal, organic soybean meal, lysine, choline and vitamin A were spiked with all 3 viruses and placed in a trailer of a commercial transport vehicle. The samples were stored in vented containers to allow exposure to environmental factors including temperature and relative humidity during the trip. Overall, the journey involved 21 days, 107 hours of transport, crossed 14 states and covered approximately 6000 miles. At the end of the study period, samples were tested for the presence of viral genome by PCR and viable virus by swine bioassay. Results indicated the presence of viable PRRSV, SVA and PEDV in both soy products, while viable SVA was recovered from all 5 ingredients. In contrast, survival was limited in the vitamins and amino acid ingredients. These results validate published laboratory data indicating the protective nature of soy-based feed ingredients and demonstrate that certain viruses, such as SVA are very stable in feed. In closing, this novel approach did demonstrate that three significant viral pathogens of pigs could survive in select feed ingredients during an actual shipping event, involving diverse environmental conditions and realistic transit periods. It is hoped that the information derived from this study will help to unify opinions across the swine industry, the veterinary profession, and governmental agencies regarding the risk of feed.
Project #: 16-262 | Principal Investigator: Rodger Main | Institutions: Iowa State University Veterinary Diagnostic Laboratory, Clemson University, Kansas State University Veterinary Diagnostic Laboratory, South Dakota State Animal Disease Research and Diagnostic Laboratory, University of Minnesota Veterinary Diagnostic Laboratory, USDA NAHLN | Posted: 8/13/2018 | Keywords: data, diagnostic, HL7, LOINC, NAHLN, standard, swine, VDL
Seamless integration of diagnostic data from any number of veterinary diagnostic laboratories (VDLs) into third-party database applications for further analytical and reporting purposes has long been recognized as a critical element necessary for proficient detection, monitoring, response, and/or management of significant diseases across a region, state, or nation. Establishing and adopting the use of universally recognized diagnostic data standards and a common electronic messaging schema are foundational elements necessary for the development of sustainable and scalable systems of connectivity and web-based analytical tools necessary to support the current needs and future demands of the US Pork Industry.
This highly collaborative initiative served to further develop the core infrastructure and diagnostic data standards necessary for US Pork Industry stakeholders to effectively harness the capabilities that any number of existing or yet to be developed (web-based) animal health information management technologies aim to provide. Primary deliverables included the development of a more comprehensive electronic message schema; a greatly expanded formulary of now more than 700 standardized test result codes covering the full-spectrum of diagnostic tests commonly conducted on swine case submissions to veterinary diagnostic labs (VDLs); an intuitive, web-based search engine that enables users (e.g., VDL information technology personnel) to readily identify the appropriate diagnostic test result code(s) (i.e., Logical Observation Identifier Names and Codes, or LOINC®) to use for each of the specific assays conducted and reported from their laboratory; and a web-based electronic message validator that allows VDLs to test, trouble-shoot, and validate their electronic messaging capabilities.
Each of the VDLs collaborating on this infrastructure development project demonstrated their ability to electronically synthesize and successfully deliver results from diagnostic case submissions utilizing the updated electronic messaging schema and expanded formulary of diagnostic test result codes. To ensure national level scalability, the updated electronic messaging schema derived from this project is consistent and fits within the overarching architecture of the Health Level Seven® (HL7) messaging schema adopted by the United States Department of Agriculture National Animal Health Laboratory Network (USDA NAHLN). The expanded formulary of standardized codes for diagnostic test results (LOINCs); the updated and more comprehensive HL7 electronic messaging schema; the standardized diagnostic test result code (LOINC®) search engine; and the HL7 electronic message validator application developed as a result of this project will be made available for use by VDLs across the USDA NAHLN.
The infrastructure developed in accordance with the primary deliverables of this initiative will provide a lasting foundation for enhancing the adoption and use of universally recognized diagnostic data standards and HL7 electronic messaging capabilities in swine interest VDLs in the USDA NAHLN. Furthering the establishment and use of such standards are critical building blocks for enhancing the connectivity and interoperability of diagnostic information across VDLs and for broadly transcending swine diagnostic information management into the digital era. Albeit these developments centered at the four VDLs collaborating on this project and were specifically focused on diagnostic tests conducted on US swine, this precedent-setting work could readily be expanded and emulated for use across any number of animal species and VDLs.
Project #: 18-137 | Principal Investigator: Diego G. Diel | Institution: South Dakota State University | Posted: 3/2/2020 | Keywords: feed biosecurity, mitigation, feed additives
Feed biosecurity became a topic of much interest to the swine industry, given recent results suggesting that feed can harbor viable viral pathogens and potentially serve as source of infection to susceptible pigs. The goal of this study was to evaluate the mitigation potential of chemical feed additives following natural consumption of contaminated and mitigated feed. To determine whether chemical mitigation of feed could reduce or prevent pathogen transmission through feed, we performed a feed trial experiment in which animals were allowed to ingest contaminated or contaminated and mitigated feed for three consecutive days. After feeding, each animal was sampled individually and levels of viremia, virus shedding and viral load in tissues were determined by RT-qPCR. For this in vivo trial we selected three candidate mitigants, A, B, and C, from our previous in vitro mitigation project. Results of our trial show that only mitigant A reduced Senecavirus A transmission through feed. Whereas no significant differences between control non-mitigated and mitigated feed were observed for porcine epidemic diarrhea virus. Results here, under conditions in which each animal ingested contaminated and mitigated feed, show that that chemical mitigation alone (with mitigants A, B, and C) may not be able to prevent transmission of pathogens through feed. These findings can be likely attributed to many factors, including: 1) the dose of virus used in our trial; 2) the fact that the chemical mitigants don’t reduce viral load in feed to levels that are non-infectious to susceptible pigs; and 3) poor contact time of the mitigant with the virus. Therefore, alternative strategies such as storage time and importation of feed ingredients from known and trusted sources should also be carefully considered to safeguard the US swine industry from unwanted viral pathogens that are endemic in other regions of the world. Additionally, studies on the mechanism of action of potential mitigants may also allow selection of those compounds that present the greatest chance of virus inactivation in the feed matrix.
Project #: 18-215 | Principal Investigator: Luis Giménez-Lirola | Institution: Iowa State University | Posted: 3/2021 | Keywords: qPCR, oral fluid, pseudorabies virus, data standardization
This research represents a collaborative effort between researchers at the Canadian Food Inspection Agency (CFIA), USDA Agricultural Research Service (ARS), and Iowa State University.
Background Beginning in the 1960s, clinical PRV became increasingly problematic in commercial swine herds in Europe, the Americas, and Southeast Asia, with annual losses estimated at $21 to $25 million (USD) to U.S. producers (Miller et al., 1996; Neumann et al., 2005). The U.S. officially eliminated PRV from domestic swine in 2004, but PRV is occasionally introduced into “transitional” herds via contact with feral swine, e.g., Minnesota (2002) and Wisconsin (2007).
Ultimately, PRV control and elimination was achieved by exploiting the molecular biology of the virus in a PRV DIVA (differentiation of infected from vaccinated animals) strategy (Quint et al., 1987; van Oirschot et al., 1986). In brief, the PRV viral envelope includes up to 11 glycoproteins (gB, gC, gD, gE, gG, gH, gI, gK, gL, gM, gN) (Mettenleiter, 2000). Relevant to PRV control, glycoprotein gB is highly conserved and required for both viral replication and cell-to-cell spread of the virus. In contrast, gE is not required for PRV replication and is associated with virulence. Wild-type viruses contain both gB and gE, while PRV MLV vaccines contain gB, but not gE. Thus, the genetic profile of vaccine virus is gB+/gE- while wild-type virus is positive for both genes (gB+/gE+). This clear distinction between wild-type and MLV viruses has allowed for the development of a DIVA approach to PRV control and eradication. That is, gene-deleted vaccines are used to protect animals against clinical PRV, with differential antibody ELISAs or DNA PCRs used to detect infected animals.
In 2018, PRV was listed as #4 in the SHIC swine disease matrix because of the potential for introducing highly pathogenic PRV into the US from Asia (https://www.swinehealth.org/swine-disease-matrix/) and its potential negative impact on exports. For this reason, improvements in PRV diagnostics, surveillance, control, and elimination remain relevant.
Project objectives The PRV PCRs have been developed for individual animal nasal swabs, but not for swine oral fluid. Therefore, the objective of this project was to evaluate the detection of PRV in swine oral fluid collected from vaccinated and/or inoculated pigs using two contemporary real-time PCR assays targeting PRV gB and gE genes.
Detection of PRV in oral fluids and nasal swabs Diagnostic samples of precisely known PRV infection status were used to evaluate assays and establish shedding dynamics. In this project, nasal swabs and oral fluid samples were collected from forty 12- to 16-week-old pigs in 4 treatment groups (10 pigs per group): PRV vaccinated with MLV and challenged with classical wild-type virus at 20 days post vaccination (DPV) (Group 1), PRV vaccinated (Group 2), wild-type PRV inoculated (Group 3), and negative control (Group 4). To detect the presence of PRV DNA, samples were tested using 1) gB PCR for screening PRV-positive animals and 2) gE triplex PCR for differentiation between PRV classic and PRV high-pathogenic (China) strains.
Results PRV DNA was detected by 0 DPV and 1 DPI in oral fluid and nasal swab samples from vaccinated and/or wild-type virus inoculated animals using the gB screening PCR. The detection of vaccine and wild-type viruses were up to 2 and 30 days, respectively. The post-inoculation detection rate was lower with shorter shedding period in vaccinated pigs (Group 1). The shedding patterns in oral fluid were comparable to which in nasal swabs. Four false positive results were observed in nasal swabs from vaccinated pigs (Group 1 and 2). The gE triplex PCR likewise detected classical strain PRV in both oral fluid and nasal swab samples from inoculated pigs (Group 3), but at a lower frequency than the gB PCR.
Conclusions This study showed that PRV DNA could be detected in swine oral fluid specimens using PRV gB and gE real-time PCRs. Furthermore, comparisons of detection rates in nasal swab vs oral fluid samples suggested that oral fluid could be used as an alternative to individual pig (nasal swab) sampling for PRV surveillance. However, further improvements in the performance of both the gB and gE PCRs would be recommended.
Contact info: Dr. Luis Gimenez-Lirola, Iowa State University, College of Veterinary Medicine, 1800 Christensen Drive, Ames, IA. Email: [email protected]. Phone: 1-515-294-7025
Project #: 18-146 | Principal Investigator: Diego G. Diel | Institution: Cornell University | Posted: 1/15/2020
The swine acute diarrhea syndrome coronavirus (SADS-CoV) RT-PCR assay was incorporated into the EZ-PED/TGE/PDCoV MPX 1.1 RT-PCR assay. The SADS RT-PCR assay replaced the TGE RT-PCR assay in the multiplex. A comparison was completed between EZ-PED/TGE/PDCoV MPX 1.1 vs EZ-PED/SADS/PDCoV MPZ. Swine fecal and oral fluid samples were randomly selected and extracted for testing with both assays side-by-side. As shown in the report, results between the two assays was very similar, indicating that the newly developed SADS-CoV is compatible and does not interfere with the PED and PDCoV assays.
The new multiplex real-time PCR (EZ-PED/SADS/PDCoV) was evaluated in China using samples with known enteric and SADS coronavirus status. This portion of the project was carried out in the laboratory of Dr. Shao Lun Zhai at the Guangdong Academy of Agricultural Sciences. All the samples tested with the new multiplex PCR had been previously tested with an in-house PCR developed at Dr. Zhai’s laboratory. As shown in the report, overall results from Dr Zhai’s PCR and the newly developed assay here are consistent, with the newly developed assay showing increased capability of detecting a few additional positive samples for all three pathogens (PEDV, PoDV and SADS-CoV) when compared to the in-house PCR. An interesting observation of these data is the fact that most SADS-CoV positive samples were also positive for PEDV or PDCoV. Some samples were also positive for all three pathogens.
Project #: 18-213 | Principal Investigator: Cesar A Corzo | Institution: University of Minnesota | Posted: – | Keywords: monitoring, emerging, PRRS, PEDV, trends
Objective 1: Monitor trends in pathogens incidence and prevalence – PRRSv, PEDv, PDCoV, Senecavirus and central nervous system associated viruses continue to be monitored in time and space. The 2018-2019 season fortunately ended with the second lowest PRRSv breeding herd cumulative incidence (21.3%) during the last 10 years of monitoring. Still, herds that break remain in category 1 for longer periods. PEDv also followed the same trend and maintained a low level of cumulative incidence with the few cases being reported throughout the country. PDCoV continues to be present in our industry clustered in specific regions; however, incidence is low as cases of this disease are not reported frequently.
Objective 2: To conduct prospective monitoring of PRRSv sequence evolution and impact – PRRSv sequence capturing process and analysis continues and our database currently has 30,603 sequences from 31 systems. Preliminary analysis showed that PRRSv sequences cluster in time and space as expected. Similarity analysis was conducted to understand how viruses have changed when compared to the reference strain VR2332. Majority of viruses are 10-15% different while there is a subset of viruses that lie between 2-5%. A similar analysis was conducted for viruses that have been classified as 1-7-4 as these viruses by comparing them to the first reported virus of this RFLP group. Diversity among newly reported 174 viruses appear to be decreasing through time together with the occurrence of this virus.
Objective 3: Develop capacity to capture and analyze movement data – The new implemented technology has been generating truck movement data consistently. A total of 12 vehicles (8 trucks and 4 trailers) are being monitored in one of the SHMP participating systems. Data structure and processes for downloading data are still being understood and procedures developed to facilitate the data analysis. A first attempt was made and transport networks were being built to understand truck movement and frequency. As expected, the data shows that movements between sow farms-growing pig sites-trucks washes are the most frequent.
Objective 4: To expand participation of producers to allow for all to be involved – Expansion continues as 2 new production companies joined the project for a total of 38. The search for new potential participants will continue. As the SQL database has enable the capability to expand in the number of farms to be added to our database, the addition of growing pig sites from specific participants is in process as we have received authorization to move forward.
Project #: 18-198 | Principal Investigator: J Zimmerman, C Wang, R Main | Institution: Iowa State University | Posted: 1/7/2020 | Keywords: FAD, spatiotemporal, spatially balanced sampling, surveillance, statistics
Effective surveillance should efficiently collect data for production and/or business planning, document freedom from specific pathogens, and guide a rapid, effective response to emerging and/or FADs. Current on-farm or regional surveillance programs routinely fail to meet these targets. In part, this is because the industry has changed over time and no longer conforms to the assumptions under which our surveillance systems were originally designed. As a result, surveillance either is not done or is done ineffectively.
Project #: 16-273 and 19-154 | Principal Investigator: Gustavo S. Silva | Institution: Iowa State University | Posted: 2/9/2021 | Keywords: biosecurity, vulnerability, scoring, risk, swine, breeding herd, PRRS
Investments in biosecurity practices are made by producers to reduce the likelihood of introducing pathogens such as porcine reproductive and respiratory syndrome virus (PRRSv). The assessment of biosecurity practices in breeding herds is usually done through surveys. The objective of this study was to evaluate the use of machinelearning (ML) algorithms to identify key biosecurity practices and factors associated with breeding herds selfreporting (yes or no) a PRRS outbreak in the past 5 years. In addition, we explored the use of the positive predictive value (PPV) of these models as an indicator of risk for PRRSv introduction by comparing PPV and the frequency of PRRS outbreaks reported by the herds in the last 5 years. Data from a case control study that assessed biosecurity practices and factors using a survey in 84 breeding herds in U.S. from 14 production systems were used. Two methods were developed, method A identified 20 variables and accurately classified farms that had reported a PRRS outbreak in the previous 5 years 76% of the time. Method B identified six variables which 5 of these had already been selected by model A, although model B outperformed the former model with an accuracy of 80%. Selected variables were related to the frequency of risk events in the farm, swine density around the farm, farm characteristics, and operational connections to other farms. The PPVs for methods A and B were highly correlated to the frequency of PRRSv outbreaks reported by the farms in the last 5 years (Pearson r=0.71 and 0.77, respectively). Our proposed methodology has the potential to facilitate producer’s and veterinarian’s decisions while enhancing biosecurity, benchmarking key biosecurity practices and factors, identifying sites at relatively higher risk of PRRSv introduction to better manage the risk of pathogen introduction.
Project #: 18-204 | Principal Investigators: Cassandra Jones, et. al. | Institution: Kansas State University | Posted: 10/2019 | Keywords: porcine, epidemic, diarrhea, virus, feed, ingredient, sampling, sensitivity, bulk
Animal feed and ingredients have the potential to transmit swine viruses. Viruses may not be evenly distributed throughout a batch of ingredients or feed, so it is difficult to properly collect a representative sample for analysis, similar to the problem of detecting aflatoxin in a load of corn. The purpose of this experiment was to determine the best type of sample to collect to determine if an ingredient is contaminated with porcine epidemic diarrhea virus (PEDV). To address this objective, we first contaminated soybean meal with two levels of PEDV (Low: 103 TCID50 per gram vs. High: 105 TCID50 per gram) or confirmed it to be PEDV negative. One hundred grams of PEDV-negative soybean meal was added to each of 13 1 kilogram polyethylene tote bags. Next, three grams of PEDV-negative soybean meal (one replicate), low PEDV soybean meal (six replicates), or high PEDV soybean meal (six replicates) was added to the corner of each tote. Finally, 887 grams of PEDV-negative soybean meal was added to each tote, bringing the final mass to 1,000 grams. A cotton gauze swab was wiped across the exterior and top of the polyethylene material. Ten individual samples were collected from each tote using aseptic probes. One gram of each probe sample was reserved for analysis, while one gram from each of the 10 probes per tote were combined to make a composite sample representing each tote. This resulted in each tote having one environmental swab, 10 probe samples, and one composite sample, which were analyzed for PEDV via qRT-PCR. As expected, no PEDV was detected from the swab, probe, or composite sample of the control samples. At the low dose, no PEDV was detected in swabs, individual probes, or composite samples, but was confirmed in 100% of the inoculant samples. At the high dose, only 37% of the probes and 33% (39.2) of the swabs had detectable PEDV. In summary, sampling bulk feed or ingredients for PEDV should include compositing at least 10 individual samples. Clearly, additional research is needed to better detect PEDV in soybean meal and environmental probes when there are low levels of contamination. Therefore, future research efforts should identify alternative methods that have a similar sensitivity to detecting PEDV, but require less time and effort to collect such a sample.
Project #: 17-189 | Principal Investigator: Megan Niederwerder, DVM, PhD | Institution: Kansas State University | Posted: 7/14/2020 | Keywords: African swine fever virus, animal feed, anti-infective agents, ASFV, domestic pig, food additives, ships, swine diseases, virus inactivation
Published Results Updated 7/14/2020 | Initial Publication
African swine fever (ASF) is currently considered the most significant global threat to pork production worldwide. Disease caused by the ASF virus (ASFV) results in high case fatality of pigs. Importantly, ASF is a trade-limiting disease with substantial implications on both global pork and agricultural feed commodities. ASFV is transmissible through natural consumption of contaminated swine feed and is broadly stable across a wide range of commonly imported feed ingredients and conditions. The objective of the current study was to investigate the efficacy of medium-chain fatty acid and formaldehyde-based feed additives in inactivating ASFV. Feed additives were tested in cell culture and in feed ingredients under a transoceanic shipment model. Both chemical additives reduced ASFV infectivity in a dose-dependent manner. This study provides evidence that chemical feed additives may potentially serve as mitigants for reducing the risk of ASFV introduction and transmission through
feed.
Project #: 18-197 | Principal Investigators: K. Poonsuk, DVM PhD, J. Zimmerman, DVM PhD, L. Giménez-Lirola, PhD | Collaborators: C. Nfon, DVM PhD, A. Clavijo, DVM PhD (National Centres for Animal Diseases – Canada) | Institution: Iowa State University
Foot-and-mouth disease virus (FMDV) remains uncontrolled in most of the world, with circulation of multiple serotypes in endemic areas. Actually, North America is among the few “FMDV-free without vaccination” areas of the world. The current massive level of global trade and traffic means that FADs anywhere in the world present a credible risk to U.S. agriculture. Our recent experience with PEDV is witness to that fact.
Preparing an effective response to the introduction of FMDV is the responsible thing to do. In the event of an FMDV outbreak in North America, effective control and elimination will require rapid detection. In turn, rapid detection will rely on an (1) efficient surveillance sampling technology and (2) immediate access to accurate diagnostic assays. Therefore, the long-term objective of this project is to create a FMD 3ABC antibody indirect ELISA (iELISA) for use with swine oral fluids.
In this study, prototype serum and oral fluid FMDV 3ABC ELISAs were developed using samples from animals of precisely known FMDV status. The optimized tests detected specific antibody in serum and oral fluid samples from FMDV-infected or FMDV-vaccinated pigs by 7-14 days post exposure. Importantly, (1) the response is not serotype specific, i.e., the 3ABC ELISAs detect antibody in animals exposed to serotypes O, A, SAT2, and Asia 1 and (2) the assays detects antibody against a non-structural protein which is not present in FMDV inactivated vaccines, i.e., the test provides for differentiation of vaccinated vs infected animals (DIVA).
Diagnostic testing of swine oral fluid samples has proven to be an effective and reliable method for the surveillance of endemic infectious diseases. Expanding this methodology to include FMDV will help provide FMDV-infected countries a new tool to control the infection and prepare the U.S. industry for a “worst-case” scenario.
Project #: 18-189 | Principal Investigator: Rodger Main |Co-investigators: Pam Zaabel, Kerry Leedom-Larson, James Roth, and Jeff Zimmerman | Institution: Iowa State University | Posted: 6/10/2019
A study was commissioned in 2018 with the aim of seeking a more in-depth understanding of the National Poultry Improvement Plan (NPIP) and assessing the potential for an NPIP like program to support the US pork industry. NPIP is a unique industry, state, and federal partnership that has long served to safeguard, improve, and assure the health of US poultry and enhance the competitiveness of the US poultry and egg industries in the domestic and global marketplace. Participation in NPIP is voluntary and almost universal among commercial poultry and egg operations throughout the US. Participants utilize NPIP to certify the health status of US poultry and egg flocks, hatcheries, slaughter plants, products, and states in accordance with NPIP’s officially recognized standards and definitions. NPIP’s health status certifications are used to demonstrate evidence of freedom of both trade and non-trade impacting diseases of poultry. NPIP is a working and active system of animal health control whose programs content and direction are informed and updated every two years by a formal congress of industry stakeholders and subject matter experts. NPIP’s program definitions, standards, and health status classifications are broadly recognized across all 50 states and by international trading partners. While participation in NPIP is voluntary, specified NPIP health status certifications are commonly required at points of sale, exhibition, and for interstate and international commerce. NPIP’s Avian Influenza Virus (AIV) surveillance programs and health status certifications held by meat-type chicken and turkey slaughter plants, commercial table egg laying operations, and states have played a primary role in helping sustain export markets and interstate commerce from unaffected regions during times of an AIV outbreak of significance affecting US commercial poultry operations.
Project #: 18-136 | Principal Investigator: Bailey Arruda | Institution: Iowa State University
Atypical porcine pestivirus (APPV) is the most common cause of congenital tremor (CT) in pigs. CT is a disease of neonatal pigs that is characterized by bilateral, clonic contractions of skeletal muscle that are seen within hours of birth and stop when piglets are at rest. APPV is transmitted from the dam to fetuses resulting in all or a subset of piglets with CT. Gilt and low parity sows are frequently reported to have CT litters; however, CT litters can be seen with multiparous sows as well. Outbreaks of CT litters, reports of up to 50% of litters affected, are most commonly observed in herds that are newly stocked, have changed genetics or have a high gilt replacement rate. In such outbreaks, it is not uncommon to have litter mortality reported to be 30 to 40%. Regardless of parity, CT litters are likely due to insufficient dam immunity at a critical timeframe of gestation. It is not currently known the most common route of exposure and infection in dams. Semen as well as cohorts may be involved.
There is limited information concerning the ecology, epidemiology and pathophysiology of APPV. Currently, there is no available serologic assay to evaluate the immunity of a dam or herd. Such an assay would provide meaningful information to assess the effectiveness of preventative measures such as acclimation and vaccination as well as improve our understating of the infection dynamics and herd impact of APPV.
To address this need, a panel of specimens including serum and oral fluid was generated that contained polyclonal antibodies against APPV. To achieve this objective, twenty two cesarean derived colostrum deprived (CDCD), crossbred, mixed-sex, 6-week-old pigs were individually identified, blocked by litter and randomly assigned to one of two groups in separate rooms based on inoculum and pen. Two pigs were placed per pen with three pens (negative control animals) or eight pens (positive control animals) per room. Oral fluids, serum, and nasal swab samples were collected prior to challenge and submitted for detection APPV by PCR prior to inoculation. Pigs were inoculated with MEM (n=6; negative control) or APPV (n=16) intravenously, intramuscularly and intranasally. Serum, pen-based oral fluids samples (3 or 8 pens/group), and individual nasal swabs were collected 0, 3, 7, 10, 14, 17, 21, 28, 35, 42, 49, 56, 63, and 70 days post inoculation (DPI). Oral fluids were also collected 31, 38, 45, 52, 59 and 66 DPI.
APPV was first detected at 10 DPI in the serum of four inoculated animals. By 35 DPI, APPV was detected in the serum of every APPV-inoculated animal and all animals remained positive until the end of the study at 70 DPI. APPV was detected in 63% and 83% of oral fluids at 14 and 17 DPI, respectively. By 24 DPI, all oral fluid samples were positive and remained positive until the end of the study at low Cq values (17.3-20.0). APPV was first detected in nasal swabs from four animals at 17 DPI with nasal shedding detected in animals at 42, 49, and 56 DPI. APPV was not detected in any sample type from negative control pigs.
With the panel of known status specimens generated at the completion of the first objective, two Erns iELISA assays (serum and oral fluid) were developed using a selected immunogenic region of Erns which was cloned, expressed and purified as a recombinant polypeptide. The serum and oral fluid assays have a 100% diagnostic specificity at a cut-off ≥0.10 and 0.15, respectively.
This is the first study to experimentally infect swine with APPV and monitor the infection dynamics overtime out to 70 DPI. The results of this experimental inoculation provide evidence that APPV viremia can be prolonged, at least 60 days could be expected following intentional exposure. These finding may be applicable if gilt acclimation protocols are undertaken. Additionally, based on the results of this study it appears that oral fluid is an appropriate and likely highly sensitive sample type to monitor herd status by RT-qPCR. Lastly, both oral fluid and serum iELISA assays can be used to evaluate individual and herd status prior to and following intervention strategies. This project provided important foundational knowledge concerning the infection dynamics of APPV in experimentally infected swine while also providing the necessary samples to develop and evaluate serologic assays that will assist in furthering our understanding APPV and preventing CT litters.
Project #: 16-271 | Investigators: Jianqiang Zhang, Charles Nfon, Chuan-Fu Tsai, Chien-Hsien Lee, Lindsay Fredericks, Qi Chen, Avanti Sinha, Sarah Bade, Karen Harmon, Pablo Piñeyro, Phillip Gauger, Yun-Long Tsai, Hwa-Tang Thomas Wang and Pei-Yu Alison Lee | Institution: BMC Veterinary Research | Posted: 6/2019 | Keywords: Senecavirus, SVA, real-time RT-PCR, insulated isothermal PCR, RT-iiPCR, POCKIT(TM)
Senecavirus A (SVA) has emerged in multiple countries in recent years. SVA infection can cause vesicular lesions clinically indistinguishable from those caused by other vesicular disease viruses, such as foot-and-mouth disease virus (FMDV), swine vesicular disease virus (SVDV), vesicular stomatitis virus (VSV), and vesicular exanthema of swine virus (VESV). Sensitive and specific RT-PCR assays for the SVA detection is necessary for differential diagnosis. Real-time RT-PCR (rRT-PCR) has been used for the detection of many RNA viruses. The insulated isothermal PCR (iiPCR) on a portable POCKIT™ device is user friendly for on-site pathogen detection. In the present study, SVA rRT-PCR and RT-iiPCR were developed and validated.
Project #: 18-193 | Investigator: Cesar A Corzo | Institution: University of Minnesota | Posted: 5/29/2019 | Keywords: swine, breeding stock, transport, biosecurity, international
Pig transport within and between countries continue to play an important role for production companies. These companies are always seeking the highest level of genetic makeup on their market pigs aiming at improving whole system economics and maintaining their competitiveness; therefore, investments into obtaining the highest genetic level involve import/export of breeding-stock animals. However, transport of livestock around the globe may represent risk as infected animals may harbor exotic pathogens that could be introduced into another country. Data on the frequency, quantity and type of pigs entering and exiting the United States is scarce. In addition, the process by which these exports/imports occurs is not well known; therefore, further understanding of the frequency of these events and procedures are warranted in order to understand whether these are risky events.
Objectives: 1) Determine the frequency of international breeding-stock exports or imports and their country of destination and/or origin; and, 2) To characterize the procedures currently implemented by breeding-stock companies during the export or import processes.
Methods: For objective 1, databases containing information related to breeding-stock imports/exports were obtained for analysis. More specifically, data sources were divided in two main groups, public databases and breeding-stock private databases. Public databases were identified through website searches whereas private databases were requested directly from individual breeding-stock companies by an invitation to participate in this project. For objective 2, participating breeding-stock companies were asked to share their transport protocols together with the possibility of us witnessing their import/export process.
Results: Based on official USDA records, between 2007 and 2018, a total of 839,152 pigs (e.g gilts or boars) were imported into the United States. Most of these pigs originated from Canada, while less than 3% were imported from Western Europe. On the other hand, breeding pig exports accounted for 382,118 pigs between 2007 and 2018 with Asia being the main destination (54.0%), followed by Mexico (31.3%) and South America (5.7%). A total of 8 breeding-stock companies were invited to participate in this study, 50% of these accepted the invitation by sharing import/export protocols. Biosecurity procedures are across companies are similar which assure the maintenance of the health status of these pigs.
Implications: The results of this study show that exports/imports are a frequent event. Imports from outside North America occurs less frequently.
Project #: 17-199 | Investigator: Douglas Marthaler | Institution: Kansas State University
Belonging to the family Picornaviridae, Porcine teschovirus (PTV) was first identified in pigs exhibiting various symptoms including ataxia, nystagmus, convulsions, polioencephalomyelitis, and paralysis. While previous molecular methods are available to detect PTV, the lack of pan-PTV ELISA (ability to detect all the serotypes) hampers our ability to conduct PTV surveillance and determination of the immune status within a herd. The aimed of our study was to develop and validate a pan-PTV indirect ELISA for diagnostics.
The VP1 protein of picornaviruses stimulates the strongest immune response but is also serotype specific. Thus, we created three protein constructs VP1, VP2, and VP2-VP1 linker. The VP2-VP1 linker protein generated the highest fluorescence levels for detection. The diagnostic sensitivity and specificity was determined using the known PTV antibody status of 558 serum samples (369 positive and 189 negatives). A cutoff of 0.4105 was used, yielding a diagnostic sensitivity and specificity of 99.2% and 98.4%, respectively. Using a single lot of internal control serum, PTV ELISA exhibited a within-plate Coefficient of Variation (CV) of 7.32%, within-run CV of 7.85%, and between-runs CV of 9.24%, indicating the highly repeatable with serum samples.
The indirect PTV ELISA was employed to investigate the immune response in post-weaned piglets after PTV exposure. During the first week post-weaning, the PTV antibody response was negative. On day 35, the ELISA detected PTV antibodies, and the detection of antibodies peaked on the last day of the study (63 days). Fluorescent microsphere immunoassay (FMIA) on the BioRad system was used to detect PTV antibodies in oral fluids. However, the ability to detect PTV antibodies in oral fluids was highly variable. Between days 35 and 41, the ability to detect PTV antibodies was sporadic, and PTV antibodies were not detected after 41 days of age. These results suggest oral fluids may not be a suitable specimen to determine the PTV immune status of a herd.
In conclusion, we developed and validated an indirect PTV ELISA for use in diagnostics. In addition, we measure the antibody response to PTV in weaned piglets and determined the PTV antibodies in oral fluids was variable.
Project #: 17-141 | Principal Investigator: Jeff Zimmerman | Institution: Iowa State University | Posted: 5/9/2019 | Keywords: emerging, foreign animal disease, spatiotemporal, spatially balanced sampling, surveillance, statistics
Effective surveillance should efficiently collect data for production and/or business planning, document freedom from specific pathogens, and guide a rapid, effective response to emerging and/or FADs. Current on-farm or regional surveillance programs routinely fail to meet these targets. In part, this is because the industry has changed over time and no longer conforms to the assumptions under which our surveillance systems were originally designed. As a result, surveillance either is not done or is done ineffectively.
On-farm surveillance The statistical theory on which on-farm surveillance was originally based assumes: (1) subjects (pigs) are independent, (2) all pigs have an equal probability of being selected for sampling, and (3) the farm has a stable, homogenous pig population. Traditional farms fit these assumptions – hence the “30 sample” approach worked in the PRV eradication program – but current swine production systems do not.
Contemporary production systems differ from traditional farms in ways that are incompatible with traditional surveillance: (1) Today’s production systems are much larger than in the past. Iowa farms averaged a total inventory of 250 animals in 1980 (Flora et al., 2007) versus 3,265 according to a study commissioned by the Iowa Pork Producers Association in 2016 (https://www.iowapork.org/study-iowa-pork-industry-remains-important-economic-driver/). (2) Pigs no longer run free in pastures or feedlots. Instead, management of large swine populations requires physical segregation by age and stage into buildings and pens. (3) Swine populations on modern farms experience rapid turnover of animals and frequent introduction of new animals – often of a different disease status. Thus, current production systems rely on extensive movement of pigs, people, trucks, and feedstuffs between sites. This connects distant places/populations and facilitates the rapid movement of pathogens between them.
Surveillance at the farm level In NPB #13-157 (Rotolo et al., 2017), we showed that disease on contemporary farms moved in a spatiotemporal fashion (non-random). This led us to develop new surveillance guidelines for on-farm surveillance based on spatial (non-random) sampling. This “fixed spatial sampling” approach is being used in the U.S. and elsewhere.
Surveillance at a regional level Efficient regional surveillance is fundamental to detecting the incursion of new pathogens and in monitoring regional disease control/elimination projects. Thus, the current project moved surveillance to the regional level with the objective of developing more efficient regional surveillance methods (fewer samples, but better detection). In this project, we tested the hypothesis that disease exhibited a spatiotemporal pattern of spread at the regional level (just as we saw on farms). The emergence of PEDV in April 2013 provided the opportunity to examine this question.
Using PEDV testing results from the Iowa State University Veterinary Diagnostic Laboratory (at the county level to protect client confidentiality), we found a spatiotemporal pattern of PEDV spread. This means that, just as for on-farm sampling, the assumptions upon which regional surveillance have been based do not hold in today’s world. This is important because it means that new guidelines for regional surveillance should be developed using statistically-appropriate modelling to account for the spatial and temporal correlation in disease spread. As a first effort in developing new guidelines, we have shown that spatially balanced sampling through generalized random–tessellation stratified (GRTS) gives a higher power of detection than traditional simple random sampling (SRS) using simulation studies mimicking real PEDV data.
Thus, our research has provided a better understanding of the spatiotemporal nature of disease spread. Initial assessment showed that use of a spatially balanced sampling scheme improved the power of disease detection and the efficiency of the disease surveillance.
Project #: 19-147 | Principal Investigator: Derald J. Holtkamp | Institutions: Iowa State University | Posted: 5/1/2019 | Keywords: swine contaminated livestock trailers, market load-outs, biosecurity, transportation
Currently, many livestock trailers in the United States are not washed, disinfected or dried between loads of market pigs due to the lack of trailers, truck washes and other swine transport related infrastructure. If livestock trailers or other carrying agents associated with the marketing event become contaminated, it is unlikely that the contamination is mitigated unless specific procedures, such as washing are performed. Under these circumstances, the livestock trailer, truck and driver returning directly from a swine slaughter plant are likely frequently contaminated with live infectious PRRSv or PEDv or both when they enter a growing pig site to haul the next load.
Project #: 18-211 | Investigator: Diego G. Diel | Institution: South Dakota State University
This study evaluated the stability of Senecavirus A, a picornavirus surrogate for foot-and-mouth disease virus (FMDV), and a pathogen that is known to survive for prolonged time in several swine feed ingredients. Common swine feed ingredients including conventional soybean meal (SBM-C), DDGS, lysine and Vitamin D were inoculated with a constant dose of SVA and incubated under different temperatures (4oC [39.6oF], 15oC [59oF] and 30oC [86oF]) to assess the effect of temperature on the stability of the virus. Samples incubated at each temperature were collected weekly for 14 weeks (days 1 through 91) and the amount of viable SVA was determined by virus titrations in the laboratory. Control samples consisted of stock virus incubated in a plastic container without a feed ingredient. The control samples were included in all temperatures tested, collected and processed following the same sample schedule as above. SVA was inactivated within 7-14 days when incubated at 30oC (86oF). Lower incubation temperatures (4oC [39.6oF], 15oC [59oF]), however, favored survival of SVA for 28 or up to 91 days, respectively. The results from this study demonstrate that SBM and DDGS provide a good matrix for the survival of SVA. Lysine and vitamin D, on the other hand only supported SVA survival for 21 days, even when incubated at lower more favorable temperatures (4oC [39.6oF]). A clear effect of temperature on the stability of the SVA was also observed. When SVA spiked-SBM or -DDGS were incubated at 4oC, infectious SVA was recovered from these samples until the end of the experiment on week 14 or day 91 post-incubation. It is important to point out that SVA viability decayed much faster (7-21 days) in control samples, in which the virus stock was deposited directly in a plastic tube without a feed matrix. The half-life, or the time required for infectious SVA amounts to decrease by one-half, were also determined in all feed ingredients. These results, consistent with the decay rate, show an extended half-life for SVA in SBM and DDGS when incubated at 4oC (10.9 and 37.9 days, respectively). Incubation at higher temperatures results in rapid degradation of the virus and very short half-life’s (1.25 and 1.36 days for SBM and DDGS, for example). In conclusion, results from these studies confirm that common swine feed ingredients such as SBM and DDGS provide a good environment for virus survival, increasing the overall stability of SVA, an important swine pathogen and surrogate for FMDV to survive for long periods of time. A clear effect of temperature was observed, with higher environmental temperatures resulting in rapid virus decay even in the most favorable feed ingredients. These results may help the swine industry to devise mitigation strategies that consider holding times for feed ingredients that are imported from countries where foreign animal diseases are endemic.
Project #: 17-187 | Investigators: Diego G. Diel and Scott Dee | Institution: South Dakota State University
The North American swine industry is under constant threat of foreign animal disease (FAD) entry. The goal of this study was to identify chemical feed additives that could be used to mitigate the risk of pathogen transmission through feed. Based on the outcome of our previous feed survival study, “high-risk” combinations of viruses and ingredients were identified. Ten mitigant candidates were selected and screened against the target pathogens. “High risk” combinations of virus and ingredient that were tested include: Senecavirus A (SVA; Soybean meal, lysine, choline and vitamin D); Porcine epidemic diarrhea virus (PEDV; Soybean meal, lysine, choline and vitamin D); Porcine reproductive and respiratory syndrome virus (PRRSV; Soybean meal and DDGS); and, Bovine herpesvirus type 1 (BoHV-1 – surrogate for pseudorabies virus [PRV]; soybean meal and soy oil cake). Results from our study show that among the 10 feed additives tested, a select group of additives presented promising efficacy against target swine pathogens. Although none of the feed additives tested completely inactivated the pathogen(s), consistent reductions in viral titers were observed when a select group of mitigants was used (KANA102 and MCFA). Interestingly, these two products showed promising results for all four viruses tested (SVA, PEDV, PRRSV and BoHV-1). Another important observation of our study is the fact that both KANA102 and MCFA are based on a blend of medium chain fatty acids. In addition to MCFA based products, Activate DA a blend of organic acids and a methionine analogue was also effective against most pathogens screened in our study. These results demonstrate that a select group of feed additives have the potential to be used as chemical mitigants to reduce viral contamination levels in feed. Further studies are warranted to assess the mechanism of action of those products and to assess their efficacy following natural ingestion of contaminated and mitigated feed.
Project #: – | Investigators: Gustavo S. Silva, Luis G. Corbellini, Daniel L.C. Linharea, Kimberlee L. Baker, Derald J. Holtkamp | Institutions: Iowa State University and Universidade Federal do Rio Grande do Sul | Posted: 10/9/2018
In modern veterinary practice, disease prevention in livestock populations has become increasingly more important (Kimman et al., 2013). This change in focus includes the adoption of biosecurity practices, which are defined as “the implementation of practices that reduce the risk of disease agents being introduced and spread into a population” (Food and Agriculture Organization, 2010).
Previous studies have demonstrated the effect of biosecurity on prevention or reduction of disease incidence (Alonso et al., 2013; Amass, 2004; Hagenaars, 2008). However, evaluation of biosecurity practices on pig farms is extremely complex. Pathogens can be introduced into pig farms in different ways (Pileri and Mateu, 2016) and the effectiveness of specific biosecurity practices depends on the characteristics of the herd, characteristics of the premises, and surrounding areas and connections to other swine premises. Porcine reproductive and respiratory syndrome (PRRS) continues to be a major health challenge in U.S. herds since it was first reported in 1989 (Keffaber, 1989). While the incidence in the U.S. has declined in recent years (Morrison et al., 2015), the prevalence continues to increase over time (MSHMP, 2018) and PRRS virus (PRRSv) still causes significant economic losses worldwide (Holtkamp et al., 2013; Nathues et al., 2017). PRRSv can be transmitted between farms via different risk events including swine movements, pickup and deliveries of supplies from or to farms, people movement, contact with other animals, air and water (Otake et al., 2002; Perez et al., 2015; Zimmerman et al., 2012).
Herd-specific biosecurity assessments are useful to determine how PRRSv may be introduced in swine herds and research is needed to quantify the relative importance of specific biosecurity practices to reduce the frequency of outbreaks. Biosecurity assessments have been used to identify relevant risk factors of disease spread onto swine farms (Bottoms et al., 2013; Holtkamp et al., 2011; Laanen et al., 2013; Sternberg Lewerin et al., 2015). However, identifying the vulnerabilities to PRRSv introduction specific to a certain production system and developing a generalized score that accounts for all major risk events is an intrinsically complex process.
Given the complexity of evaluating biosecurity practices to prevent the introduction of PRRSv, applying a technique that uses multiple factors to score swine breeding herds based on their relative vulnerability to PRRSv introduction would be beneficial for prioritizing and identifying gaps in biosecurity practices and predicting the frequency of outbreaks. Several methods exist to evaluate these factors, allowing for a ranking of specific factors by relative importance. One method by which to do this is multi-criteria decision analyses (MCDA) (Belton and Stewar, 2002), which has been applied extensively in a variety of fields (Santos et al., 2017; Steele et al., 2009; Thokala et al., 2016), including to assess vulnerability (Cardona, 2003; Joerin et al., 2010). MCDA was chosen for the present study because it provides a systematic way to integrate information from a range of sources, compare scenarios and prioritize decisions (Cox et al., 2013).
The objective of this study was to develop a biosecurity vulnerability score (BVS) that represents the relative vulnerability of swine breeding herds to the introduction of PRRSv. To validate the BVS, a survey of biosecurity practices and PRRS outbreak histories in 125 breed-to-wean herds in two different populations in the U.S. was used. Data on the frequency of PRRS outbreaks was used to test the hypothesis that BVS were different between farms that have a low incidence of PRRS outbreaks, compared to farms that have a high incidence.
Project #: 17-188 | Investigator: Cassandra Jones | Institution: Kansas State University
Senecavirus A (SVA), previously known as Senecavirus A, is a detrimental pathogen in the United States swine industry. Transmission is not well understood, but its similarity to foot and mouth disease virus (FMDV) suggests direct contact with people or fomites may spread the virus. Once present, viruses in feed, feed ingredients, and feed mills are difficult to mitigate. While contaminated surfaces in a feed mill have been demonstrated as a potential vector for bacterial and viral transmission, there is currently no approved method for its evaluation of viral contamination. Therefore, the objective of this Experiment 1 was to validate standardized swabbing techniques for detection of SVA. A secondary objective was to determine if a freeze/thaw cycle impacted detectable RNA. This experiment included 3 forms (inoculum, feed, or swab), 4 doses of SVA (none, low, medium, or high), and 2 storage methods (analyzed initially vs. after a freeze/thaw cycle). The SVA was added to swine feed, with 1 g reserved, and the remaining spread over a stainless steel coupon. Feed was removed, but residual feed dust remained. Next, surfaces were swabbed and samples split, with one set analyzed initially, and another frozen for 7 days, then thawed and analyzed. Results are reported as the quantity of detectable SVA as determined by threshold cycle (Ct) in qRT-PCR, where the higher the Ct, the less detectable virus was identified. The results demonstrate that sample type impacted the quantity of detectable SVA, where feed samples were approximately 8 Ct higher than the inoculum, and swab samples were approximately 4 Ct higher than feed. A freeze/thaw cycle did not impact detectable SVA compared to samples that were analyzed immediately.
In Experiment 2, the objective was to determine the prevalence and distribution of SVA in United States swine feed mills as an indicator of risk of domestic and foreign animal disease transmission through feed. A total of 375 samples were collected from 11 surfaces + one feed sample collected from 11 different feed mills manufacturing swine feed located in 8 different states. Feed mills include 5 producing both mash and pelleted feed in KS, CO, OK, NC, and IA, and 6 producing only mash feed in KS, NC, MN, IA, IN, and IL. Within a mill, locations included ingredient pit grating, fat intake inlets, exterior of pellet mill (only in feed mills with pelleting capacity), finished product boot bin, load-out auger, finished feed, floor dust in the break/control room, floor dust in receiving, floor dust in the manufacturing area, floor dust in the warehouse, worker shoe bottoms, and broom in the manufacturing area. To account for potential seasonality associated with pathogenic hazards, the same locations in feed mills were swabbed in Late Fall 2016, Winter 2016/17, and Summer 2017. Notably, no mills were manufacturing feed for SVA-positive herds at the time of analysis. Five of 375 samples analyzed positive for SVA, with Ct ranging from 37.4 to 39.9. One positive sample was collected in late Fall, while the other four positive samples were collected in Winter. No positive samples were identified in Summer. Two samples were from load-out augers, and one each from fat intake inlet, floor dust in the receiving area, and worker shoes. A sow farm being fed by the mill with SVA on worker shoes was subsequently diagnosed with SVA after the sample as collected.
These results indicate that an environmental swab can be used to detect SVA in feed, however with approximately 4 Ct less precision than analyzing feed samples directly. Furthermore, the limit of detection of SVA in environmental swabs appears to be near 10^3 TCID50/mL. Samples can be frozen prior to analysis without impacting detectable SVA RNA. Finally, SVA was not widespread throughout the swine feed mills analyzed in this experiment, but its presence in a mill may be indicative of disease risk or entry into pig populations, particularly through worker shoes.
Project #: 17-191 | Investigators: J. Zimmerman | Institution: Iowa State University | Posted: 7/6/2018 | Keywords: foot-and-mouth disease, ELISA
Foot-and-mouth disease virus (FMDV) remains uncontrolled in most of the world, with circulation of multiple serotypes in endemic areas. Actually, North America is among the few “FMDV-free without vaccination” areas of the world. The current massive level of global trade and traffic means that FADs anywhere in the world present a credible risk to U.S. agriculture. Our recent experience with PEDV is witness to that fact.
Project #: 16-250 | Principal Investigator: Aruna Ambagala | Institution: National Centre for Foreign Animal Disease- Winnipeg, MB | Posted: 7/18/2018 | Keywords: pseudorabies, virulent, variant, detection, real-time, PCR, assay, DIVA
Pseudorabies virus (PRV) causes pseudorabies or Aujeszky’s disease in livestock and wild mammals; however pigs are the main host and reservoir for this virus. It causes deadly disease in newborn piglets, respiratory problems in growing and fattening pigs, and reproductive problems in pregnant sows. Like other herpesviruses, PRV establishes a lifelong infection in the nervous system followed by subsequent shedding of infectious virus. Pseudorabies has spread throughout the world, but Canada, Greenland, and Australia are considered free of this disease. In 2004, PRV was eliminated from the US commercial swine herds, but the virus remains in some localized feral swine populations. China is considered the largest pork producer in the world. The earliest documented PRV outbreak in China was in 1947. Since 1990s, more than 80% of pigs in China have been vaccinated and the clinical disease was well controlled. In late 2011 however, a newly PRV virus (variant) which cause severe disease surfaced in PRV vaccinated pig herds in Northern China. Since then, this virus has spread across China causing severe economic losses.
Copyright 2025 | Swinehealth.org | Website by Heartland Marketing Group