
The Swine Health Information Center, launched in 2015 with Pork Checkoff funding, protects and enhances the health of the US swine herd by minimizing the impact of emerging disease threats through preparedness, coordinated communications, global disease monitoring, analysis of swine health data, and targeted research investments.
On this episode of SHIC Talk, host Barbara Campbell Determan is joined by Foundation for Food & Agriculture Research Scientific Program Manager Dr. Miriam Martin LeValley along with SHIC Executive Director Dr. Megan Niederwerder and Associate Director Dr. Lisa Becton. They talk about the ongoing SHIC/FFAR/NPB collaboration directing H5N1 research with 10 round 1 projects awarded in summer 2025 as well as a second request for proposals issued and due March 24, 2026.
Join SHIC along with the American Association of Swine Veterinarians for a webinar on Senecavirus A as an emerging disease risk and the need for diligence to differentiate from foot-and-mouth disease virus, a foreign animal disease of concern to US pork producers. Expert presenters will provide the latest information on the current SVA situation in domestic pig herds, research updates, and insights into disease investigations. The webinar will be conducted by the Swine Medicine Education Center at Iowa State University.
March 26, 2026
1:00 – 2:30 pm CT

Each year, the Swine Health Information Center updates its Plan of Work developed from stakeholder priorities. The 2026 Plan of Work, which guides SHIC’s activities, contains 25 projects and topics designed to fulfill SHIC’s five strategic priorities: 1) improve swine health information, 2) monitor and mitigate risks to swine health, 3) responding to emerging disease, 4) surveillance and discovery of emerging disease, and 5) swine disease matrices. SHIC’s 2025 Progress Report contains details on the record return on investment for the Center’s activities to protect and enhance the health of the US swine herd, based on Plan of Work deliverables.
Industry stakeholders provide input into the development of the Plan of Work which is then approved by the SHIC Board of Directors. Activities directed by the 2026 Plan of Work will be implemented by Executive Director Dr. Megan Niederwerder and Associate Director Dr. Lisa Becton with input from the board and SHIC Working Groups.
Primary funding for SHIC’s 2026 Plan of Work comes from the Pork Checkoff under a contract between both organizations. In 2026, the National Pork Board voted to provide $1.5 million to fund SHIC. SHIC’s 2026 Plan of Work reflects this budget while maintaining its focus on deliverables to the US swine industry and the pork producers who fund the Checkoff.
While the 2026 Plan of Work directs activities for SHIC, the organization is nimble and able to respond to industry needs as they arise. Stakeholder input and ideas are welcomed year-round to inform newly identified industry needs which may necessitate adapting the Plan of Work to fulfill SHIC’s mission. A request for proposals to address 2026 Plan of Work priorities is planned for release later this year.
In 2026, SHIC continues its ongoing partnership with the Foundation for Food & Agriculture Research and Pork Checkoff aiming to fill knowledge gaps regarding H5N1 Risk to Swine through a collaborative research program. Ten H5N1 projects on swine were funded in 2025 to address gaps in knowledge identified through producer input. In January 2026, SHIC released a second round of request for proposals to fulfill remaining H5N1 research priorities.
SHIC 2026 Plan of Work Priorities
Improve Swine Health Information
Monitor and Mitigate Risks to Swine Health
Responding to Emerging Disease
Surveillance and Discovery of Emerging Disease
Swine Disease Matrix
▪️Using the bacterial and viral swine disease matrices as guidelines for research to enhance swine disease diagnostic capabilities

A Swine Health Information Center-funded study aimed to improve detection of common and emerging swine pathogens using a next generation sequencing panel. Led by Dr. Rebecca Wilkes at Purdue University, the study sought to develop and validate a targeted next-generation sequencing (tNGS) respiratory panel capable of detecting multiple viral and bacterial swine respiratory pathogens in a single test. Swine respiratory disease is often caused by multiple pathogen infections occurring simultaneously, making diagnosis and control difficult when relying on single-pathogen tests. Results from this study showed strong agreement between tNGS and PCR, high specificity, and robust sensitivity across clinically relevant pathogen loads. Moreover, the panel successfully identified mixed infections and detected pathogens that may be missed by targeted PCR testing alone.
Find the industry summary for SHIC project #24-007 here.
Infectious disease syndromes in swine are frequently multifactorial and can be challenging to determine the causative pathogen(s) by clinical signs alone. Co-infections with viral and bacterial pathogens often occur, making accurate diagnoses challenging. Many diagnostic tests utilized in swine are single pathogen assays. Consequently, there is a critical need within the swine industry for comprehensive diagnostic approaches capable of simultaneously detecting co-infections and identifying emerging or unexpected pathogens. This comprehensive approach can help support more informed treatment decisions, guide targeted interventions, and reduce reliance on sequential or repeated testing.
Objectives of the study described herein include 1) develop and optimize primers targeting swine respiratory pathogens, 2) validate specificity and sequencing performance with characterized strains, 3) determine analytical sensitivity in comparison with qPCR and 4) validate diagnostic performance using clinical samples. An array of primers targeting multiple regions across the genomes of common and emerging swine respiratory pathogens was designed in collaboration with the AgriSeq Bioinformatics team (Thermo Fisher Scientific, USA). The panel was developed to enable simultaneous detection of viral, bacterial, and selected virulence-associated targets relevant to swine disease syndromes. Primer design focused on highly conserved genomic regions to ensure broad detection, while selected targets included regions enabling genotyping or lineage assignment for specific pathogens, such as PRRSV and PCV2.
Selected clinical samples were retrospectively obtained from swine cases submitted to the Purdue University Animal Disease Diagnostic Laboratory (ADDL) based on results from the laboratory’s validated real-time PCR assays. Pathogens included PRRSV, Influenza A virus, PCV2, PCV3, M. hyopneumoniae (MHP), Streptococcus suis, Glaesserella parasuis, Actinobacillus pleuropneumoniae (APP), and M. hyorhinis (MHR). Clinical isolates of Streptococcus suis were also included. Sample selection was performed to ensure representation of both viral and bacterial respiratory pathogens as well as mixed infections commonly associated with porcine respiratory disease complex.
A total of 70 clinical samples were included in this study, including 20 positive samples to establish relative limits of detection. An additional 25 PCR-positive respiratory cases and 25 PCR-negative respiratory case controls were used to determine diagnostic sensitivity, specificity and accuracy of the developed test. Feasibility of the targeted sequencing assay was evaluated using a combination of qPCR-positive clinical specimens, and synthetic DNA controls (gBlocks, IDT, Coralville, IA) representing targeted genomic regions.
A comprehensive multiplex primer pool targeting approximately 25 swine respiratory pathogens was successfully developed in collaboration with Thermo Fisher Scientific’s AgriSeq bioinformatics team. Successful amplification and sequencing were achieved for the respiratory pathogens included in the assay, except Actinobacillus suis, for which no positive clinical sample was available to confirm the panel’s ability to detect this pathogen. Assessment of analytical sensitivity across multiple dilutions showed the tNGS assay consistently detected major respiratory pathogens, including PRRSV (multiple lineages), PCV2, PCV3, and M. hyopneumoniae, demonstrating robust performance in complex clinical matrices.
Across the clinical validation cohort, the tNGS panel showed strong concordance with PCR testing, reflected by an overall agreement of 92% and a Cohen’s kappa value of 0.84, indicating near-perfect categorical agreement beyond chance. Importantly, all PCR-negative samples remained negative by tNGS, underscoring the assay’s high specificity and low risk of false-positive reporting. This is a critical consideration for diagnostic laboratories, where false positives can lead to unnecessary interventions, economic loss, and erosion of producer confidence.
Discrepancies between methods were limited to PCR-positive samples and were consistently associated with targets detected at high Ct values, indicating low pathogen abundance. Some targets, specifically IAV and respiratory coronavirus, performed below expectations. For the RNA viruses, this is likely due to lack of adequate primers to detect sequence differences. Future redesign of primers should improve the detection of these pathogens. Notably, no instances were observed in which tNGS detected respiratory pathogens in samples deemed negative by PCR, reinforcing the conservative and clinically appropriate nature of the assay’s detection criteria.
Evaluation of analytical sensitivity using serially diluted clinical samples confirmed that the tNGS assay retains robust detection capability across a wide range of pathogen concentrations, including those commonly encountered in routine diagnostic submissions. Beyond simple presence or absence, the tNGS panel was designed to include genotype and strain, informative genomic regions for selected viral pathogens of interest. This design enabled additional genetic resolution for several viruses, including differentiation of PRRSV North American (Type 2) versus European (Type 1) genotypes, identification of PCV2 genotype D versus non-D genotypes, and determination of influenza A virus subtypes when SIV was detected.
However, full lineage level discrimination was not consistently achieved for all targets, particularly for PRRSV in clinical specimens. PRRSV results were conservatively reported at the genotype level rather than at the lineage level. These findings indicate that, while the current assay reliably detects PRRSV presence, additional optimization or redesign of lineage-targeting primers may be required to consistently achieve lineage-level resolution, which was an intended objective of the panel design.
The sequence level data generated by the tNGS approach supports improved interpretation of circulating viral diversity and may aid outbreak investigations and epidemiologic assessments when sufficient sequence coverage is achieved. Equally important is the assay’s compatibility with herd level sample types, including pooled and population representative specimens. This expands its utility from individual case diagnosis to broader monitoring programs, where understanding pathogen diversity and circulation patterns is often more informative than identifying a single agent in a single animal.
Overall, this project demonstrates that a multiplex tNGS assay can function as a reliable and informative diagnostic tool for swine respiratory disease, with performance closely aligned to established PCR testing while offering substantially broader pathogen coverage and genomic insight. Further, tNGS can help overcome inherent limitations of targeted PCR testing alone. The study supports the use of tNGS as a complementary diagnostic and surveillance tool that can improve outbreak investigations, inform vaccine strategies, strengthen swine health monitoring programs, and expand pathogen surveillance capacity for emerging diseases.

In an article published by National Hog Farmer on February 3, 2026, Dr. Michele Moncrief, a post-doctoral research associate with the Swine Medicine Education Center at Iowa State University, detailed a recently completed study on telehealth-based biosecurity hazard analysis. Funded by the Iowa Pork Producers Association, the study examined the use of telehealth technologies as an option for expanding biosecurity evaluation capacity without increasing on-farm traffic and employed the Swine Health Information Center-funded Standardized Outbreak Investigation Program.
Designed to help veterinarians and producers identify and prioritize risks for pathogen entry, the SHIC-funded SOIP enables a consistent approach across individual users and farms. Goals of the SOIP output are to prevent disease introduction and prepare for seasonal challenges so that production systems can enhance biosecurity control measures accordingly.
Dr. Moncrief and team used the SOIP rather than developing a new assessment tool. “SOIP offers a structured, systematic framework for identifying and evaluating biosecurity hazards,” she stated. “We chose to build upon an established, field-validated program that is already recognized and used within the swine industry.” Using SOIP allowed the ISU team to align methods with real-world investigations, a process made easier due to their familiarity with the investigation platform. Dr. Moncrief described SOIP as a reliable foundation for evaluating how telehealth-based methods could be applied to biosecurity hazard analysis.
The structure offered by SOIP breaks biosecurity risk into clearly defined categories: entry events, operational procedures, and site characteristics. “This makes it well-suited for evaluating new technologies like telehealth. Because the framework already organizes observations in a consistent and repeatable way, it allowed us to directly compare findings from traditional on-site evaluations with remote, telehealth-based assessments,” Dr. Moncrief explained. “This consistency helped ensure that the differences we observed were related to the evaluation method itself rather than changes in how hazards were assessed.”
During the study, Dr. Moncrief said remote reviewers recognized many of the same risks observed during on-site analyses. “Agreement was strongest for clearly observable infrastructure and procedures, while more complex, multi-step practices were more challenging to evaluate remotely and depended on the completeness of the video footage,” she remarked. “Overall, the results suggest that SOIP supports comparable assessments across delivery methods when supported by standardized data collection and training.”
And those results speak directly to the study findings. As Dr. Moncrief wrote in the National Hog Farmer article, “Overall, remote evaluators identified many of the same biosecurity hazards as the on-site investigator. Across more than 4,200 question-level comparisons, agreement between in-person and telehealth-based evaluations averaged about 63%. In practical terms, this means that telehealth captured a meaningful portion of the biosecurity picture, but not all of it.”
Reaching this conclusion was made possible because the team used the standardized question set. “Using SOIP’s standardized question set ensured that every evaluator assessed the same biosecurity risks using the same criteria, allowing us to directly compare responses between in-person and telehealth evaluations. This made it possible to quantify agreement objectively and clearly identify which types of hazards, particularly more complex procedures, were harder to assess remotely, something that would have been much more difficult with a non-standardized review,” Dr. Moncrief said. The tool eliminated differences in evaluation approach and interpretation, which would have complicated methodological differences.
“SOIP’s successful adaptation in this study suggests that SOIP is a flexible and dynamic tool that can be used not only during outbreaks but also proactively, before a disease event occurs. The framework helps veterinarians and producers systematically identify potential gaps in biosecurity and think through disease entry risks in a structured, logical way,” Dr. Moncrief observed. “Using SOIP outside of an emergency setting allows operations to make incremental improvements and targeted reinvestments in biosecurity without the urgency and pressure that typically accompany an active outbreak.”
“It’s structured framework allows veterinarians, producers, and even external evaluators to assess risks consistently and track biosecurity improvements over time. Applying SOIP in these settings can support more objective evaluations and help operations strengthen preparedness before a disease event occurs,” Dr. Moncrief stated.
In a large-scale disease event where rapid capacity expansion is required, SOIP can play a significant role. Dr. Moncrief observed, “SOIP can support consistency by giving investigators a shared structure, terminology, and standardized question set, allowing multiple evaluators, whether on-site or remote, to assess hazards using the same framework. In our study, this helped remote reviewers identify many of the same risks as the on-site investigator, although results also showed that an orientation to the tool would likely improve alignment among evaluators, particularly in remote settings.”
The SMEC at ISU study also revealed SOIP’s usability. “Anecdotally, production managers and veterinarians who participated across multiple sites appeared to become comfortable using the framework quickly; while this was not formally assessed, it suggested the tool may be intuitive once applied in practice,” Dr. Moncrief observed.
The telehealth-based biosecurity hazard analysis project as well as results of other on-farm investigations using SOIP demonstrate the program’s value. “Investing in tools like SOIP before a crisis occurs helps ensure the industry is prepared rather than reacting under pressure when a disease event happens. Developing and validating these frameworks in advance allows veterinarians and producers to have practical, tested resources ready when rapid decision-making is required. Proactive investment by industry-supported organizations helps strengthen preparedness, reduce response time, and ultimately protects animal health and business continuity when challenges arise,” Dr. Moncrief said.
The study led by Dr. Moncrief and colleagues showed that telehealth-based hazard analysis using the framework was feasible and allowed remote evaluators to identify many of the same biosecurity risks observed during in-person assessments, not as a replacement but as a complementary component of the evaluation process. She said the process highlights how investing in standardized tools ahead of time creates opportunities to adapt and explore novel approaches as industry needs evolve.
“From this study, the most important takeaway is that a standardized framework like SOIP provides a consistent way to evaluate biosecurity risks, even when assessments are conducted by different evaluators or through different delivery methods. Our findings suggest that this shared structure helps maintain alignment in how hazards are identified and interpreted, which becomes especially important when capacity needs to expand during a disease event. That consistency builds confidence in the decisions being made, which, in turn, supports maintaining business continuity during disruptions,” Dr. Moncrief concluded.
The development of resources that support continued utilization of the SOIP helps SHIC fulfill its mission to protect and enhance the health of the US swine herd by supporting efforts to prevent, respond to and mitigate emerging, re-emerging, and transboundary swine diseases.
A recent study, funded by the Swine Health Information Center and led by Drs. Cesar Corzo and Marcello Melini at the University of Minnesota, explored the use of tongue tip samples as a viable option for effective disease diagnosis in growing pigs. The industry summary for the SHIC project #23-063 can be found here. Inspired by this project to continue exploring alternative sample types that can be used for swine diagnostics, Drs. Corzo and Melini completed another study to investigate cardiac puncture (CP) blood collection as a practical and biosecure method for post-mortem pathogen detection in pigs. Published in January 2026, the full study is now available in Frontiers in Veterinary Science.
In routine farm settings, sample collection often depends on ease, staff expertise, and the suspected pathogen, yet it must always be guided by a clear diagnostic purpose. Through investigating alternative sample types, researchers sought to identify appropriate, efficient, and easy-to-obtain samples from swine mortalities for pathogen detection. Additionally, project goals included identifying sample types that minimize the risk of environmental contamination which can lead to disease transmission as compared to traditional sample collection through necropsy or tongue-tip fluids.
Researchers investigated post-mortem CP as a minimally invasive, animal-welfare friendly and biosecure alternative that has shown promise for detecting PRRSV and could enhance diagnostic safety during endemic, emerging and foreign animal disease investigations. In the published study report, authors wrote, “This method was not only feasible for obtaining blood and testing the sera, but also avoided environmental contamination with blood, offering an alternative tool for collection during outbreak investigations.”
For this project, blood samples were collected at five Midwestern US farms, three of which were breed-to-wean operations and two of which were growing pig farms undergoing a PRRSV outbreak. To perform the study, researchers collected a total of 286 CP blood samples originating from 196 suckling and 90 growing pigs. Reported outcomes included nine cases where sample collection did not yield enough serum volume for testing, and four instances where the PCR reaction was inhibited. Among the remaining samples, PRRSV was detected in 95% of samples with a median cycle threshold value of 21.5 (Q1 17.1; Q3 28.5), a minimum of 10.8, and a maximum of 35.5.
CP offers a practical and safe alternative for specimen collection during infectious disease investigations in swine. While collection of pre-mortem blood samples remains a preferred diagnostic sample type due to its reliability, post-mortem CP provides an efficient way to obtain blood for pathogen detection without the extensive tissue disruption associated with necropsy. The study notes that necropsy-based sampling, though common for assessing lesions and collecting tissues for histology or molecular testing, often results in blood spillage. This can increase the risk of environmental contamination—a particular concern for highly stable viruses such as African swine fever virus, which can persist in blood for extended periods.
This sampling method should serve as a complementary tool during outbreak investigations, since CP is not meant to substitute for a full necropsy, which remains essential for confirming diagnoses in clinically affected pigs. The study summary states, “Blood collection from dead pigs is a viable welfare-friendly alternative for PRRSV detection. After training to collect this specimen, CP is an alternative to obtaining a blood sample for surveillance and diagnosis of pathogens of interest (i.e., PRRSV, ASF, CSF) while minimizing blood spillage and environmental contamination, which can increase the risk of pathogen dissemination.”
The authors remarked, “Our results highlight the feasibility of obtaining blood samples from recently dead pigs of different ages through CP for viral disease (i.e., PRRSV) diagnosis and surveillance. Obtaining this sample was not only possible but welfare-friendly and avoided blood spillage and thus environmental contamination, supporting our efforts for disease containment.” Investigation into alternative sample types for use in disease outbreak response and emerging disease detection provides additional tools for veterinarians and helps SHIC fulfill its mission to protect the health of US swine herds.
Reference:
Melini CM, Kikuti M, Yue X and Corzo CA (2026). Cardiac puncture blood collection as a practical and biosecure method for post-mortem pathogen detection in pigs. Front. Vet. Sci. 12:1741832. doi: 10.3389/fvets.2025.1741832.

Information sharing ranks as one of the Swine Health Information Center’s key priorities. Using a variety of mediums and methods, SHIC disseminates swine disease news, research results, disease monitoring information, and more. In addition to a highly visited website (www.swinehealth.org), SHIC shares content through its monthly newsletter, webinars on timely topics, podcasts, media interviews, social media, and presentations. In 2025, SHIC was asked to provide a weekly animal health update to WHO Radio, Des Moines, Iowa, for their Saturday farm programming.
SHIC Executive Director Dr. Megan Niederwerder and Associate Director Dr. Lisa Becton record 4.5-minute segments for the WHO Radio program. In addition to being heard on WHO’s AM signal at 1040, programming is streamed at www.iHeart.com, making this a new tool to share timely swine health information wherever the streaming signal is available.
Since June 2025, more than 30 SHIC animal health updates have been recorded. You can find these listed by date and topic on SHIC’s website here.
Swine industry stakeholders are encouraged to listen to these reports and share the information with those who may find it useful.
SHIC offers its thanks to Bob Quinn and Duane Murley, farm broadcasters at WHO Radio, for the invitation and for sharing SHIC’s message on a new frontier.

This month’s Domestic Swine Disease Monitoring Report highlights several key findings. After PRRSV reached peak case positivity in wean-to-market in October 2025, it is now in its third month with decreased detection; however, detection in sow farms continued to increase for the fifth consecutive month. PEDV case positivity continues to increase and is above expected levels for both adult/sow farms and wean-to-market. The bonus page highlights an eight-year milestone in the development of the SDRS and its contributions to the industry. The accompanying podcast features Mark Schwartz, director of innovation and production optimization at Schwartz Farms, who discusses regional and field-level clinical impact of PRRSV 1C.5.32, approaches to integrating biosecurity with control and elimination strategies for MHP and PEDV, and reflects on eight years of SDRS and its role in strengthening decision-making across the U.S. swine industry.

In the March report, read about African swine fever case escalation in South Korea where 20 outbreaks were confirmed across seven provinces by mid-February. ASF eradication has been declared in Saxony (Germany), after 2,398 wild boar cases and five and a half years of control efforts. No new detections were reported in the region for one year, meeting regional eradication criteria. In Spain, the ASF high-risk zone was expanded due to wild boar cases, prompting restriction expansion in Catalonia; no domestic pig infections have been reported. A foot-and-mouth disease outbreak in Cyprus results in the Larnaca district being placed under quarantine after confirmation in cattle, with epidemiological links to sheep and goat holdings. Learn more about surveillance efforts at points of entry in the United Kingdom (Dover & Harwich) where officials seized a record 34 tonnes of illegal meat and nearly 300 kg of illegal pork in January amid ASF and FMD biosecurity concerns.
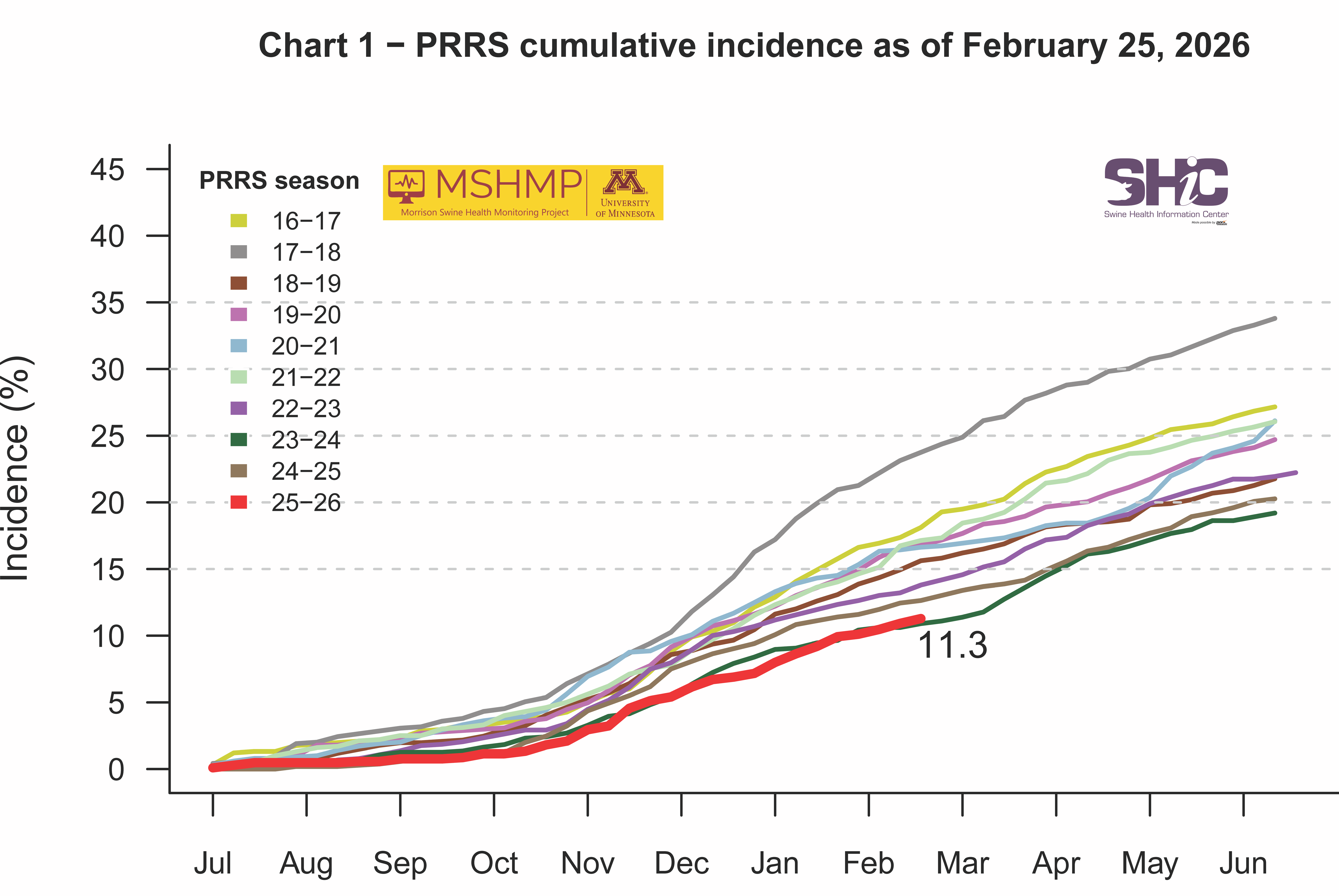
PRRS Cumulative Incidence for MSHMP
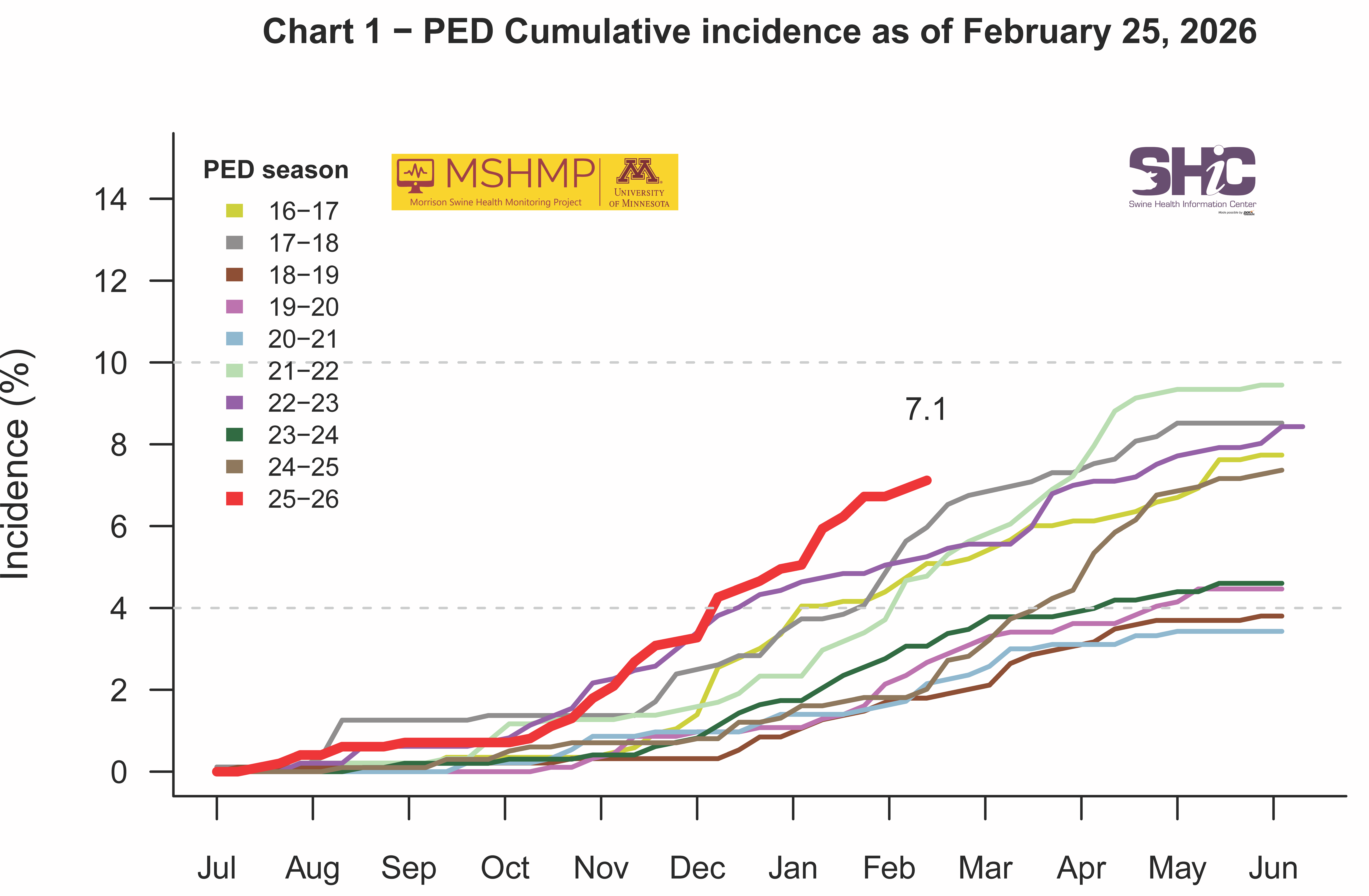
PEDV Cumulative Incidence for MSHMP
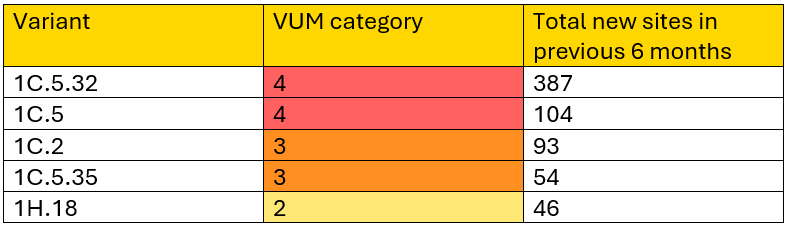
Five PRRSV variants are classified as Variants Under Monitoring (VUM) Category 2 or higher in this month’s report. Variants 1C.5 and 1C.5.35 were promoted to VUM Categories 4 and 3, respectively. Variants 1C.5.32 and 1H.18 remain at VUM Categories 4 and 2, respectively, while Variant 1C.5.33 was demoted to VUM Category 1. These changes are reflected in new variant-specific situation reports). Previous reports for all variants ever classified as VUM Category 2 or higher remain available.